Abstract
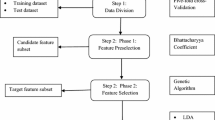
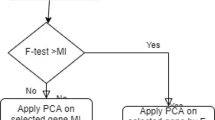
Neuromuscular disorder is a complex progressive health problem which results in muscle weakness and fatigue. In recent years, with emergence and development of machine learning- and sequencing-driven technologies, the prediction of neuromuscular disorders could be made on the basis of gene expression for accurate diagnosis of disease. The intent is to correctly distinguish the patients affected from neuromuscular disorder from the healthy one with the help of various classification methods used in machine learning. In this paper, we proposed a novel feature selection method which applies deep learning method for grouping the outputs generated through various classifiers. The feature selection is performed on the basis of integrated Bhattacharya coefficient and genetic algorithm (GA) where fitness is computed on the basis of ensemble outputs of various classifiers which is performed using deep learning methods. The Bhattacharya coefficient computed the most effective gene subset; then, the most discriminative gene subset will be formulated using GA. The proposed integrated deep learning multi-model ensemble method was applied on two commercially available neuromuscular disorder datasets. The obtained results encouraged that the proposed integrated approach enhances the prediction accuracy of neuromuscular disorders as compared with different datasets and other classifier algorithms. The proposed deep learning-driven ensemble method provides more accurate and effective results for neuromuscular disorder prediction and classification with the help of distinguished classifiers.






Similar content being viewed by others
References
Rahman S, Hanna MG (2009) Diagnosis and therapy in neuromuscular disorders: diagnosis and new treatments in mitochondrial diseases. J Neurol Neurosurg Psychiatry 80(9):943–953. https://doi.org/10.1136/jnnp.2008.158279
Power LC, O’Grady GL, Hornung TS, Jefferies C, Gusso S, Hofman PL (2018) Imaging the heart to detect cardiomyopathy in Duchenne muscular dystrophy: a review. Neuromuscul Disord 28(9):717–730
Sayed E, Wahed A, Emam IA, Badr A (2012) Feature selection for cancer classification: an SVM based approach. Int J Comput Appl 46(8):20–26
Hernandez Hernandez JC, Duval B, Hao JK (2007) A genetic embedded approach for gene selection and classification of microarray data. In: Marchiori E, Moore JH, Rajapakse JC (eds) Evolutionary computation, machine learning and data mining bioinformatics, EvoBIO 2007. Lecture Notes in Computer Science, vol. 4447. Springer, Berlin
Daliri MR (2012) Feature selection using binary particle swarm optimization and support vector machines for medical diagnosis. Biomed Tech Biomed Eng 57(5):395–402
Daliri MR (2012) Predicting the cognitive states of the subjects in functional magnetic resonance imaging signals using the combination of feature selection strategies. Brain Topogr 25(2):129–135
Wolpert DH (1992) Stacked generalization. Neural Networks 5(2):241–259
Tan AC, Gilbert D (2003) Ensemble machine learning on gene expression data for cancer classification. Appl Bioinform 2:S75–S83
Cho SB, Won HH (2003) Machine learning in dna microarray analysis for cancer classification. In Asia-Pacific bioinformatics conference, pp 189–198
Mamoshina P, Vieira A, Putin E, Zhavoronkov A (2016) Applications of deep learning in biomedicine. Mol Pharm 13:1445–1454
Gupta D, Julka A, Jain S, Aggarwal T, Khanna A, de Albuquerque VHC (2018) Optimized cuttlefish algorithm for diagnosis of Parkinson’s disease. Cognit Syst Res 52:36–48
Gupta D, Sundaram S, Khanna A, Hassanien AE, de Albuquerque VHC (2018) Improved diagnosis of Parkinson’s disease based on optimized crow search algorithm. Comput Electr Eng 68:412–424
Hira ZM, Gillies DF (2015) A review of feature selection and feature extraction methods applied on microarray data. Adv Bioinform. https://doi.org/10.1155/2015/198363
Gupta D, Rodrigues JJPC, Sundaram S, Khanna A, Korotaev V, Albuquerque VHC (2018) Usability feature extraction using modified crow search algorithm: a novel approach. Neural Comput Appl. https://doi.org/10.1007/s00521-018-3688-6
Shankar K, Lakshmanaprabu SK, Gupta D, Maseleno A, de Albuquerque VHC (2018) Optimal features-based multi kernel SVM approach for thyroid disease classification. J Supercomput. https://doi.org/10.1007/s11227-018-2469-4
Gupta A, Ahlawat A (2019) Taxonomy of GUM and usability prediction using GUM multistage fuzzy expert system. Int Arab J Inf Technol 16(3):23–39
González-Navarro FF, Belanche-Muñoz LA, Silva-Colón KA (2013) Effective classification and gene expression profiling for the facioscapulohumeral muscular dystrophy. PLoS ONE 8(12):e82071
Yao B, Li S (2010) ANMM4CBR: a case-based reasoning method for gene expression data classification. Algorithms Mol Biol 5(1):1
Tang J, Alelyani S, Liu H (2014) Feature selection for classification: a review. In: Aggarwal CC (ed) Data classification: algorithms and applications. CRC Press, Boca Raton, pp 37–52
Lakshmanaprabu SK, Shankar K, Khanna A, Gupta D, Rodrigues JJPC, Pinheiro PR, de Albuquerque VHC (2018) Effective features to classify big data using social internet of things. IEEE Access 6:24196–24204
Gupta D, Ahlawat A (2017) Usability feature selection via MBBAT: a novel approach. J Comput Sci 23:195–203
Tiwari P, Qian J, Li Q, Wang B, Gupta D, Khanna A, Rodrigues J, Albuquerque V (2018) Detection of subtype blood cells using deep learning. Cognit Syst Res 52:1036–1044
Rodrigues R, Rodrigues JJPC, Cruz M, Khanna A, Gupta D (2018) An IoT-based automated shower system for smart homes. In: International conference on advances in computing, communications and informatics (ICACCI’18)
Jain R, Gupta D, Khanna A (2018) Usability feature optimization using MWOA. In: International conference on innovative computing and communication (Proceeding of ICICC 2018, volume 2)
Daliri MR (2012) A hybrid automatic system for the diagnosis of lung cancer based on genetic algorithm and fuzzy extreme learning machines. J Med Syst 1001:5
Daliri MR (2014) A hybrid method for the decoding of spatial attention using the MEG brain signals. Biomed Signal Process Control 10:308–312
Huang W, Zeng S, Wan M, Chen G (2016) Medical media analytics via ranking and big learning: a multi-modality image-based disease severity prediction study. Neurocomputing 204:125–134
Saeys Y, Inza I, Larrañaga P (2007) A review of feature selection techniques in bioinformatics. Bioinformatics 23(19):2507–2517
Kourou K, Exarchos TP, Exarchos KP, Karamouzis MV, Fotiadis DI (2015) Machine learning applications in cancer prognosis and prediction. Comput Struct Biotechnol J 13:8–17
http://scikit-learn.org/stable. Accessed 15 Aug 2018
Silver D, Huang A, Maddison CJ, Guez A, Sifre L et al (2016) Mastering the game of go with deep neural networks and tree search. Nature 529(7587):484–489
Shin HC, Roth HR, Gao M, Lu L (2016) Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging 35(5):1285–1298
Bakay M, Wang Z, Melcon G et al (2006) Nuclear envelope dystrophies show a transcriptional fingerprint suggesting disruption of Rb–MyoD pathways in muscle regeneration. Brain 129(4):996–1013
Aherne FJ, Thacker NA, Rockett PI (1998) The Bhattacharyya metric as an absolute similarity measure for frequency coded data. Kybernetika 34(4):363–368
Babu KG, Prasad MR (2013) An effective approach in face recognition using image processing concepts. Int J Appl Innov Eng Manag 2(8):215–219
Acknowledgements
This work was supported in part by the Indian Council of Social Science Research under Grant No. 02/138/2017-18/RP/Major. The authors would like to thank the reviewers in advance for their comments and suggestions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors do not have financial and personal relationships with other people or organizations that could inappropriately influence (bias) their work.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Khamparia, A., Singh, A., Anand, D. et al. A novel deep learning-based multi-model ensemble method for the prediction of neuromuscular disorders. Neural Comput & Applic 32, 11083–11095 (2020). https://doi.org/10.1007/s00521-018-3896-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-018-3896-0