Abstract
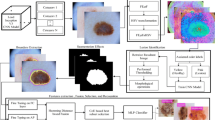
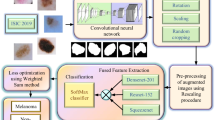
Malignant melanoma, not belongs to a common type of skin cancers but most serious because of its growth—affecting large number of people worldwide. Recent studies proclaimed that risk factors can be substantially reduced by making it almost treatable, if detected at its early stages. This timely detection and classification demand an automated system, though procedure is quite complex. In this article, a novel strategy is adopted, which not only diagnoses the skin cancer but also assigns a proper class label. The proposed technique is principally built on saliency valuation and the selection of most discriminant deep features selection. The lesion contrast is being enhanced using proposed Gaussian method, followed by color space transformation from RGB to HSV. The new color space facilitates the saliency map construction process, utilizing inner and outer disjoint windows, by making the foreground and background maximally differentiable. From the segmented images, deep features are extracted by utilizing inception CNN model on two basic output layers. These extracted set of features are later fused using proposed decision-controlled parallel fusion method, prior to feature selection using proposed window distance-controlled entropy features selection method. The most discriminant features are later subjected to classification step. To demonstrate the efficiency of the proposed methods, three freely available datasets are utilized such as PH2, ISBI 2016, and ISBI 2017 with achieve accuracy is 97.74%, 96.1%, and 97%, respectively. Simulation results clearly reveal the improved performance of proposed method on all three datasets compared to existing methods.

















Similar content being viewed by others
References
Oliveira RB, Papa JP, Pereira AS, Tavares JMR (2018) Computational methods for pigmented skin lesion classification in images: review and future trends. Neural Comput Appl 29:613–636
Khan MA, Akram T, Sharif M, Shahzad A, Aurangzeb K, Alhussein M et al (2018) An implementation of normal distribution based segmentation and entropy controlled features selection for skin lesion detection and classification. BMC Cancer 18:638
Akram T, Khan MA, Sharif M, Yasmin M (2018) Skin lesion segmentation and recognition using multichannel saliency estimation and M-SVM on selected serially fused features. J Ambient Intell Human Comput. https://doi.org/10.1007/s12652-018-1051-5
CFAFACS. https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2018/cancer-facts-and-figures-2018.pdf. Accessed May 3, 2018
Parkin D, Mesher D, Sasieni P (2011) 13. Cancers attributable to solar (ultraviolet) radiation exposure in the UK in 2010. Br J Cancer 105:S66
Nguyen AH, Detty SQ, Agrawal DK (2017) Clinical implications of high-mobility group box-1 (HMGB1) and the receptor for advanced glycation end-products (RAGE) in cutaneous malignancy: a systematic review. Anticancer Res 37:1–7
WABASCSCACS. http://www.cancer.org/cancer/skincancer-basalandsquamouscell/detailedguide/skin-cancer-basal-and-squamous-cell-what-is-basal-and-squamous-cell. Accessed Jan 31, 2018
Nasir M, Attique Khan M, Sharif M, Lali IU, Saba T, Iqbal T (2018) An improved strategy for skin lesion detection and classification using uniform segmentation and feature selection based approach. Microsc Res Tech 81:528–543
Whiteman DC, Green AC, Olsen CM (2016) The growing burden of invasive melanoma: projections of incidence rates and numbers of new cases in six susceptible populations through 2031. J Investig Dermatol 136:1161–1171
Ruela M, Barata C, Marques JS, Rozeira J (2017) A system for the detection of melanomas in dermoscopy images using shape and symmetry features. Comput Methods Biomech Biomed Eng Imaging Vis 5:127–137
Satapathy SC, Fernandes SL, Lin H (2019) Stroke lesion segmentation and analysis using entropy/Otsu’s function—a study with social group optimization. Curr Bioinform 14:305–313
Saba T, Khan MA, Rehman A, Marie-Sainte SL (2019) Region extraction and classification of skin cancer: a heterogeneous framework of deep CNN features fusion and reduction. J Med Syst 43:289
Liaqat A, Khan MA, Shah JH, Sharif M, Yasmin M, Fernandes SL (2018) Automated ulcer and bleeding classification from WCE images using multiple features fusion and selection. J Mech Med Biol 18:1850038
Fernandes SL, Tanik UJ, Rajinikanth V, Karthik KA (2019) A reliable framework for accurate brain image examination and treatment planning based on early diagnosis support for clinicians. Neural Comput Appl 29:1–12
Aurangzeb K, Haider I, Khan MA, Saba T, Javed K, Iqbal T et al (2019) Human behavior analysis based on multi-types features fusion and Von Nauman entropy based features reduction. J Med Imaging Health Inform 9:662–669
Amin J, Sharif M, Yasmin M, Fernandes SL (2018) Big data analysis for brain tumor detection: deep convolutional neural networks. Future Gener Comput Syst 87:290–297
Bokhari F, Syedia T, Sharif M, Yasmin M, Fernandes SL (2018) Fundus image segmentation and feature extraction for the detection of glaucoma: a new approach. Curr Med Imaging Rev 14:77–87
Acharya UR, Fernandes SL, WeiKoh JE, Ciaccio EJ, Fabell MKM, Tanik UJ et al (2019) Automated detection of Alzheimer’s disease using brain MRI images—a study with various feature extraction techniques. J Med Syst 43:302
Jin Z, Zhou G, Gao D, Zhang Y (2018) EEG classification using sparse Bayesian extreme learning machine for brain–computer interface. Neural Comput Appl. https://doi.org/10.1007/s00521-018-3735-3
Zhang Y, Nam CS, Zhou G, Jin J, Wang X, Cichocki A (2018) Temporally constrained sparse group spatial patterns for motor imagery BCI. IEEE Trans Cybern 49:1–11
Zhang Y, Zhang H, Chen X, Liu M, Zhu X, Lee S-W et al (2019) Strength and similarity guided group-level brain functional network construction for MCI diagnosis. Pattern Recogn 88:421–430
Jiao Y, Zhang Y, Wang Y, Wang B, Jin J, Wang X (2018) A novel multilayer correlation maximization model for improving CCA-based frequency recognition in SSVEP brain–computer interface. Int J Neural Syst 28:1750039
Sharif M, Khan MA, Faisal M, Yasmin M, Fernandes SL (2018) A framework for offline signature verification system: best features selection approach. Pattern Recognit Lett. https://doi.org/10.1016/j.patrec.2018.01.021
Fernandes SL, Chakraborty B, Gurupur VP, Prabhu G (2016) Early skin cancer detection using computer aided diagnosis techniques. J Integr Des Process Sci 20:33–43
Khan MA, Akram T, Sharif M, Saba T, Javed K, Lali IU et al (2019) Construction of saliency map and hybrid set of features for efficient segmentation and classification of skin lesion. Microsc Res Tech 82:741–763
Afza F, Khan MA, Sharif M, Rehman A (2019) Microscopic skin laceration segmentation and classification: a framework of statistical normal distribution and optimal feature selection. Microsc Res Tech. https://doi.org/10.1002/jemt.23301
Khan MA, Javed MY, Sharif M, Saba T, Rehman A (2019) Multi-model deep neural network based features extraction and optimal selection approach for skin lesion classification. In: 2019 international conference on computer and information sciences (ICCIS), pp 1–7
Chatterjee S, Dey D, Munshi S (2018) Optimal selection of features using wavelet fractal descriptors and automatic correlation bias reduction for classifying skin lesions. Biomed Signal Process Control 40:252–262
Codella NC, Nguyen Q-B, Pankanti S, Gutman D, Helba B, Halpern A et al (2017) Deep learning ensembles for melanoma recognition in dermoscopy images. IBM J Res Dev 61:5:1–5:15
Goyal M, Yap MH (2017) Multi-class semantic segmentation of skin lesions via fully convolutional networks. Preprint arXiv:1711.10449
Ross-Howe S, Tizhoosh H (2018) The effects of image pre-and post-processing, wavelet decomposition, and local binary patterns on U-nets for skin lesion segmentation. Preprint arXiv:1805.05239
Sarker M, Kamal M, Rashwan HA, Banu SF, Saleh A, Singh VK et al (2018) SLSDeep: skin lesion segmentation based on dilated residual and pyramid pooling networks. Preprint arXiv:1805.10241
Lopez AR, Giro-i-Nieto X, Burdick J, Marques O (2017) Skin lesion classification from dermoscopic images using deep learning techniques. In: 2017 13th IASTED international conference on biomedical engineering (BioMed), pp 49–54
Kawahara J, BenTaieb A, Hamarneh G (2016) Deep features to classify skin lesions. In: 2016 IEEE 13th international symposium on biomedical imaging (ISBI), pp 1397–1400
Celebi ME, Zornberg A (2014) Automated quantification of clinically significant colors in dermoscopy images and its application to skin lesion classification. IEEE Syst J 8:980–984
Chen S, Wang Z, Shi J, Liu B, Yu N (2018) A multi-task framework with feature passing module for skin lesion classification and segmentation. In: 2018 IEEE 15th international symposium on biomedical imaging (ISBI 2018), pp 1126–1129
Amin J, Sharif M, Yasmin M, Fernandes SL (2017) A distinctive approach in brain tumor detection and classification using MRI. Pattern Recognit Lett. https://doi.org/10.1016/j.patrec.2017.10.036
Klang E (2018) Deep learning and medical imaging. J Thorac Dis 10:1325
Naqi S, Sharif M, Yasmin M, Fernandes SL (2018) Lung nodule detection using polygon approximation and hybrid features from CT images. Curr Med Imaging Rev 14:108–117
Raza M, Sharif M, Yasmin M, Khan MA, Saba T, Fernandes SL (2018) Appearance based pedestrians’ gender recognition by employing stacked auto encoders in deep learning. Future Gener Comput Syst 88:28–39
Khan MA, Akram T, Sharif M, Javed MY, Muhammad N, Yasmin M (2018) An implementation of optimized framework for action classification using multilayers neural network on selected fused features. Pattern Anal Appl 22:1–21
Sharif M, Khan MA, Akram T, Javed MY, Saba T, Rehman A (2017) A framework of human detection and action recognition based on uniform segmentation and combination of Euclidean distance and joint entropy-based features selection. EURASIP J Image Video Process 2017:89
Khan MA, Sharif M, Javed MY, Akram T, Yasmin M, Saba T (2017) License number plate recognition system using entropy-based features selection approach with SVM. IET Image Proc 12:200–209
Yang J, Yang J-Y, Zhang D, Lu J-F (2003) Feature fusion: parallel strategy vs. serial strategy. Pattern Recogn 36:1369–1381
Adeel A, Khan MA, Sharif M, Azam F, Umer T, Wan S (2019) Diagnosis and recognition of grape leaf diseases: an automated system based on a novel saliency approach and canonical correlation analysis based multiple features fusion. Sustain Comput Inform Syst. https://doi.org/10.1016/j.suscom.2019.08.002
Rashid M, Khan MA, Sharif M, Raza M, Sarfraz MM, Afza F (2019) Object detection and classification: a joint selection and fusion strategy of deep convolutional neural network and SIFT point features. Multimed Tools Appl 78:15751–15777
Arshad H, Khan MA, Sharif M, Yasmin M, Javed MY (2019) Multi-level features fusion and selection for human gait recognition: an optimized framework of Bayesian model and binomial distribution. Int J Mach Learn Cybern. https://doi.org/10.1007/s13042-019-00947-0
Pathan S, Prabhu KG, Siddalingaswamy P (2018) Hair detection and lesion segmentation in dermoscopic images using domain knowledge. Med Biol Eng Comput 56:1–15
Pennisi A, Bloisi DD, Nardi D, Giampetruzzi AR, Mondino C, Facchiano A (2016) Skin lesion image segmentation using delaunay triangulation for melanoma detection. Comput Med Imaging Graph 52:89–103
Bi L, Kim J, Ahn E, Kumar A, Fulham M, Feng D (2017) Dermoscopic image segmentation via multi-stage fully convolutional networks. IEEE Trans Biomed Eng 64:2065–2074
Bozorgtabar B, Sedai S, Roy PK, Garnavi R (2017) Skin lesion segmentation using deep convolution networks guided by local unsupervised learning. IBM J Res Dev 61:6:1–6:8
Yang J, Xie F, Fan H, Jiang Z, Liu J (2018) Classification for dermoscopy images using convolutional neural networks based on region average pooling. IEEE Access 6:65130–65138
Harangi B (2018) Skin lesion classification with ensembles of deep convolutional neural networks. J Biomed Inform 86:25–32
Oliveira RB, Pereira AS, Tavares JMR (2017) Skin lesion computational diagnosis of dermoscopic images: ensemble models based on input feature manipulation. Comput Methods Programs Biomed 149:43–53
Yu L, Chen H, Dou Q, Qin J, Heng P-A (2017) Automated melanoma recognition in dermoscopy images via very deep residual networks. IEEE Trans Med Imaging 36:994–1004
Vasconcelos CN, Vasconcelos BN (2017) Experiments using deep learning for dermoscopy image analysis. Pattern Recognit Lett. https://doi.org/10.1016/j.patrec.2017.11.005
Maia LB, Lima A, Pereira RMP, Junior GB, de Almeida JDS, de Paiva AC (2018) Evaluation of melanoma diagnosis using deep features. In: 2018 25th international conference on systems, signals and image processing (IWSSIP), pp 1–4
Bi L, Kim J, Ahn E, Feng D, Fulham M (2016) Automatic melanoma detection via multi-scale lesion-biased representation and joint reverse classification. In: 2016 IEEE 13th international symposium on biomedical imaging (ISBI), pp 1055–1058
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors have no conflict of interest and contribute equally in this work for results compilation and other technical support.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Khan, M.A., Akram, T., Sharif, M. et al. An integrated framework of skin lesion detection and recognition through saliency method and optimal deep neural network features selection. Neural Comput & Applic 32, 15929–15948 (2020). https://doi.org/10.1007/s00521-019-04514-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-019-04514-0