Abstract
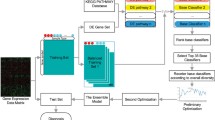
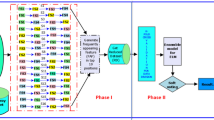
Although cancer diagnosis research has continuously made breakthroughs in a single indicator, it is a challenging task to improve its multiple joint indicators. This study proposes a multi-category multi-state information ensemble-based classification method. We fuse protein-coding and non-coding genes to construct co-expression profiles, which ensemble the field information of classical genetics and epigenetics. A hierarchical feature selection algorithm based on control groups is put forward to quickly remove irrelevant and redundant features without the bias caused by unbalanced dataset. Multiple heterogeneous diagnosis models, which ensemble multiple diagnosis model structures and model states, are constructed and a competition mechanism is then introduced to automatically select the best model from multiple heterogeneous models without deeply grasping the positive and negative fusion effects between different algorithms and features. We apply the proposed method to classify three high-incidence cancers, in which the classification accuracy and sensitivity are over 99.23% and the classification specificity is over 97.37%. This illustrates that the proposed method has upgraded the three joint indicators of cancer diagnosis at the same time. Compared with the state-of-the-art classification methods, the classification accuracy has been improved by 2.23–9.23%, the sensitivity by 6.25–37.40%, and the specificity by 0–12.02%. In addition, feature analysis reveals three biological findings.





Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Data availability
All raw data is from TCGA. The datasets used and analyzed in the current study are presented in additional supporting files.
Abbreviations
- PCGs:
-
Protein-coding genes
- NCGs:
-
Non-coding genes
- PCG-EPs:
-
Protein-coding gene expression profiles
- NCG-EPs:
-
Non-coding gene expression profiles
- CO-EPs:
-
Co-expression profiles
- C-EPs:
-
Cancer expression profiles
- CG:
-
Comparison group
- HFSA:
-
Hierarchical feature selection algorithm
- SAMM:
-
Single algorithm & multi-model
- MASM:
-
Multi-algorithm & single model
- MAMM:
-
Multi-algorithm & multi-model
- MHDMs:
-
Multiple heterogeneous diagnosis models
- MMIECM:
-
Multi-category multi-state information ensemble-based classification method
References
Jemal A, Siegel R, Xu J et al (2010) Cancer statistics, 2010. CA-Cancer J Clin 63(1):11. https://doi.org/10.3322/caac.21166
Laura J, Hongyue D, Marc J et al (2002) Gene expression profiling predicts clinical outcome of breast cancer. Nature 415(6871):530–536. https://doi.org/10.1038/415530a
Wang M, Klevebring D, Lindberg J, Czene K, Grönberg H, Rantalainen M (2016) Determining breast cancer histological grade from RNA-sequencing data. Breast Cancer Res 1:48. https://doi.org/10.1186/s13058-016-0710-8
Salem H, Attiya G, El-Fishawy N (2016) Classification of human cancer diseases by gene expression profiles. Appl Soft Comput 50:124–134. https://doi.org/10.1016/j.asoc.2016.11.026
Wang Y, Wang D, Geng N, Wang Y, Yin Y, Jin Y (2019) Stacking-based ensemble learning of decision trees for interpretable prostate cancer detection. Appl Soft Comput 77:188–204. https://doi.org/10.1016/j.asoc.2019.01.015
Nguyen T, Khosravi A, Creighton D, Nahavandi S (2015) Hidden markov models for cancer classification using gene expression profiles. Inform Sci 316:293–307. https://doi.org/10.1016/j.ins.2015.04.012
Esteller M (2011) Non-coding RNAs in human disease. Nat Rev Genet 12(12):861–874. https://doi.org/10.1038/nrg3074
Lu J, Getz G, Miska E et al (2005) MicroRNA expression profiles classify human cancers. Nature 435:834–838. https://doi.org/10.1038/nature03702
Luo JW, Pan C, Xiang G, Yin Y (2019) A novel cluster-based computational method to identify miRNA regulatory modules. Ieee Acm T Comput Bi 16:681–687. https://doi.org/10.1109/Tcbb.2018.2824805
Cheerla N, Gevaert O (2017) MicroRNA based pan-cancer diagnosis and treatment recommendation. BMC Bioinformatics 18:1–11. https://doi.org/10.1186/s12859-016-1421-y
Saha I, Bhowmick S, Geraci F, Pellegrini M, Bhattacharjee D et al (2015) Analysis of next-generation sequencing data of miRNA for the prediction of breast cancer. Lect Notes Comput Sci (including Subser Lect Notes Swarm, Evolutionary, and Memetic Computing) 9873:116–127. https://doi.org/10.1007/978-3-319-48959-9_11
Zhang W, Huang J, Chen HN et al (2020) A cancer diagnosis method combining miRNA-lncRNA interaction pairs and class weight competition. IEEE Access 8:67059–67074. https://doi.org/10.1109/access.2020.2985405
Saeys Y, Inza I, Larranaga P (2007) A review of feature selection techniques in bioinformatics. Bioinformatics 23(19):2507–2517. https://doi.org/10.1093/bioinformatics/btm344
Huerta E, Montiel A, Caporale R, Lopez MA (2016) Hybrid framework using multiple-filters and an embedded approach for an efficient selection and classification of microarray data. IEEE ACM T Comput Bi 13(1):12–26. https://doi.org/10.1109/TCBB.2015.2474384
Pérez-Rodríguez J, de Haro-Garcia A, del Castillo J et al (2018) A general framework for boosting feature subset selection algorithms. Inform Fusion 44:147–175. https://doi.org/10.1016/j.inffus.2014.10.005
Kar S, Das Sharma K, Maitra M (2015) Gene selection from microarray gene expression data for classification of cancer subgroups employing PSO and adaptive k-nearest neighborhood technique. Expert Syst Appl 42(1):612–627. https://doi.org/10.1016/j.eswa.2014.08.014
Cao J, Zhang L, Wang BJ, Li FZ, Yang JW (2015) A fast gene selection method for multi-cancer classification using multiple support vector data description. J Biomed Inform 53:381–389. https://doi.org/10.1016/j.jbi.2014.12.009
Wang A, An N, Chen G, Li L, Alterovitz G (2015) Accelerating wrapper-based feature selection with k-nearest-neighbor. Knowl-Based Syst 83:81–91. https://doi.org/10.1016/j.knosys.2015.03.009
Tian Y, Sun M, Deng Z, Luo J, Li Y (2017) A new fuzzy set and nonkernel SVM approach for mislabeled binary classification with applications. IEEE T Fuzzy Syst 25(6):1536–1545. https://doi.org/10.1109/TFUZZ.2017.2752138
Maldonado S, López J (2018) Dealing with high-dimensional class-imbalanced datasets: Embedded feature selection for SVM classification. Appl Soft Comput 67:94–105. https://doi.org/10.1016/j.asoc.2018.02.051
Murata T, Yanagisawa T, Kurihara T, Kaneko M, Jinno H (2019) Salivary metabolomics with alternative decision tree-based machine learning methods for breast cancer discrimination. Breast Cancer Res Tr 177(3):591–601. https://doi.org/10.1007/s10549-019-05330-9
Moorthy K, Mohamad MS (2012) Random forest for gene selection and microarray data classification. Bioinformation 7(3):142–146. https://doi.org/10.6026/97320630007142
Wang ST, Wang YY, Wang DJ, Yin YQ, Wang YZ, Jin YC (2020) An improved random forest-based rule extraction method for breast cancer diagnosis. Appl Soft Comput 86:105941. https://doi.org/10.1016/j.asoc.2019.105941
Liu KH, Zeng ZH, Ng VTY (2016) A hierarchical ensemble of ECOC for cancer classification based on multi-class microarray data. Inform Sciences 349–350:102–118. https://doi.org/10.1016/j.ins.2016.02.028
Nagarajan R, Upreti M (2017) An ensemble predictive modeling framework for breast cancer classification. Methods 131:128–134. https://doi.org/10.1016/j.ymeth.2017.07.011
Zhou M, Jin M (2019) Holographic ensemble forecasting method for short-term power load. IEEE T Smart Grid 10(1):425–434. https://doi.org/10.1109/Tsg.2017.2743015
Tomczak K, Czerwińska P, Wiznerowicz M (2015) Review the cancer genome atlas (TCGA): an immeasurable source of knowledge. Współczesna Onkologia 1A:68–77. https://doi.org/10.5114/wo.2014.47136
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Bio. https://doi.org/10.1186/s13059-014-0550-8
Peker M (2016) A decision support system to improve medical diagnosis using a combination of k-medoids clustering based attribute weighting and SVM. J Med Syst. https://doi.org/10.1007/s10916-016-0477-6
Zhao JM, Cheng W, He XG, Liu YL, Li J et al (2018) Construction of a specific svm classifier and identification of molecular markers for lung adenocarcinoma based on lncrna-mirna-mrna network. Oncotargets Ther 11:3129–3140. https://doi.org/10.2147/OTT.S151121
Magna G, Casti P, Jayaraman SV, Salmeri M, Mencattini A et al (2016) Identification of mammography anomalies for breast cancer detection by an ensemble of classification models based on artificial immune system. Knowl-Based Syst 101:60–70. https://doi.org/10.1016/j.knosys.2016.02.019
Grail Inc (2018) Grail announces data on detection of early-stage lung cancers. Businesswire. https://www.businesswire.com/news/home/20180602005048/en/GRAIL-Announces-Data-Detection-Early-StageLung-Cancers. Accessed 02 June 2018
Ma XJ, Dahiya S, Richardson E, Erlander M, Sgroi DC (2009) Gene expression profiling of the tumor microenvironment during breast cancer progression. Breast Cancer Res. https://doi.org/10.1186/bcr2222
Acknowledgements
XianFang Tang and Zhe Shi contributed equally to this work. This work was supported in part by the National Natural Science Foundation of China under Grant 61773157, and Changsha Key R&D Program under Grant KQ2004011.
Funding
This work was supported in part by the National Natural Science Foundation of China under Grant 61773157, and in part by the Changsha Key Research and Development Program under Grant KQ2004011.
Author information
Authors and Affiliations
Contributions
XT: Editing and Submission, ZS: Software, Experiment, and Writing. MJ: Conceptualization, Methodology, Experiment Scheme, Writing, and Review.
Corresponding author
Ethics declarations
Conflicts of interest
All authors have not competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Tang, X., Shi, Z. & Jin, M. Multi-category multi-state information ensemble-based classification method for precise diagnosis of three cancers. Neural Comput & Applic 33, 15901–15917 (2021). https://doi.org/10.1007/s00521-021-06211-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-021-06211-3