Abstract
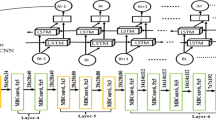
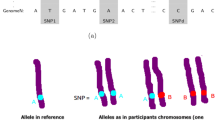
In Genome-Wide Association Studies (GWAS), detection of T2D-related variants in genome sequences and accurate modeling of the complex structure of the relevant gene are of great importance for the diagnosis of diabetes. For this purpose, this paper presents a novel strong algorithm to accurately and effectively identify Type 2 Diabetes (T2D) risk variants at high-performance rates. The proposed algorithm consists of five important phases. The first stage is to collect T2D-associated DNA sequences and to digitize them by the Entropy-based technique. The second stage is to transform these digitized DNA sequences into 224 × 224 pixels size spectrum images. The third is to extract a distinctive feature set from these spectrum images using the ResNet and VGG19 architectures. The fourth is to classify the effective feature set using SVM and k-NN methods. The last stage is to evaluate the system with k-fold cross-validation. As a result of the developed algorithm, the performances of the used Convolutional Neural Network (CNN) methods, the Entropy-based technique, and the classifiers were compared in relation. As a result of the study a combination model of the proposed Entropy-based technique, ResNet and Support Vector Machine (SVM) achieved the highest accuracy rate with 99.09%. With this study, the performance of the system in the extraction of epigenetic features and prediction of T2D from spectrogram images was investigated. The results show that the system will contribute to the identification of all genes in diabetes-related tissue and studies on new drug targets.











Similar content being viewed by others
References
Ho DSW, Schierding W, Wake M, Saffery R, O’Sullivan J (2019) Machine learning SNP based prediction for precision medicine. Front Genet. https://doi.org/10.3389/fgene.2019.00267
Imani M, Ghoreishi S, F. (2020) Optimal finite-horizon perturbation policy for inference of gene regulatory networks. IEEE Intell Syst. https://doi.org/10.1109/MIS.2020.3017155
Guariguata L, Whiting DR, Hambleton I, Beagley J, Linnenkamp U, Shaw JE (2014) Global estimates of diabetes prevalence for 2013 and projections for 2035. Diabetes Res Clin Pract 103:137–149
Arikoglu H, Kaya DE (2015) Tip 2 diyabetin moleküler genetik temeli; Son gelişmeler. Genel Tıp Dergisi 25:147–159
Defronzo RA, Ferrannini E, Groop L, Henry RR, Herman WH, Holst JJ et al (2015) Type 2 diabetes mellitus. Nat Rev Dis Primers 1:15019. https://doi.org/10.1038/nrdp.2015.19
Morris AP (2018) Progress in defining the genetic contribution to type 2 diabetes susceptibility. Curr Opin Genet Dev 50:41–51
Das KW, Elbein SC (2006) The Genetic basis of type 2 diabetes. Cell Sci 2:100–131
Mahajan A, Taliun D, Thurner M, Robertson NR, Torres JM, Rayner NW et al (2018) Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet. https://doi.org/10.1038/s41588-018-0241-6
Vinuela A, Varshney A, van de Bunt M, Prasad RB, Asplund OB, Bennett A et al (2019) Influence of genetic variants on gene expression in human pancreatic islets-implications for type 2 diabetes. BioRxiv. https://doi.org/10.1101/655670
Varshney A, Scott LJ, Welch RP, Erdos MR, Chines PS, Narisu N et al (2017) Genetic regulatory signatures underlying işlet gene expression and type 2 diabetes. Proc Natl Acad Sci 114:2301–2306. https://doi.org/10.1073/pnas.162119214
Kleinberger JW, Pollin TI (2015) Personalized medicine in diabetes mellitus: current opportunities and future prospects. Ann N Y Acad Sci 1346:45–56. https://doi.org/10.1111/nyas.12757
Awotunde JB et al (2021) Chapter Nine—Prediction and classification of diabetes mellitus using genomic data. In: Sangaiah AK, Mukhopadhyay S (eds) Intelligent IoT systems in personalized health care. Academic Press, pp 235–292
Abdulaimma B, Fergus P, Chalmers C, Montañez C (2020) Deep learning and genome-wide association studies for the classification of type 2 diabetes. In: içinde 2020 international joint conference on neural networks (IJCNN), Tem, pp 1–8. https://doi.org/10.1109/IJCNN48605.2020.9206999
Rai V et al (2020) Single-cell ATAC-Seq in human pancreatic islets and deep learning upscaling of rare cells reveals cell-specific type 2 diabetes regulatory signatures. Mol Metab 32:109–121. https://doi.org/10.1016/j.molmet.2019.12.006
Mattis KK, Gloyn LA (2020) From Genetic association to molecular mechanisms for Islet-cell dysfunction in type 2 diabetes. J Mol Biol 432:1551–1578. https://doi.org/10.1016/j.jmb.2019.12.045
Wang K, Zhou W, Meng P, Wang P, Zhou C, Yao Y, Wu S, Wang Y, Zhao J, Zou D, Jin G (2019) Immune-related somatic mutation genes are enriched in PDAGs with diabetes. Transl Oncol 12(9):1147–1154
Kumar A, JeyaSundaraSharmila D, Singh S (2017) SVMRFE based approach for prediction of most discriminatory gene target for type II diabetes. Genom Data 12:28–37. https://doi.org/10.1016/j.gdata.2017.02.008
Lalrohlui F, Zohmingthanga J, Hruaii V, Kumar NS (2020) Genomic profiling of mitochondrial DNA reveals novel complex gene mutations in familial type 2 diabetes mellitus individuals from Mizo ethnic population, Northeast India. Mitochondrion. https://doi.org/10.1016/j.mito.2019.12.001
Liang F et al (2020) Insulin-resistance and depression cohort data mining to identify nutraceutical related DNA methylation biomarker for type 2 diabetes. Genes Dis. https://doi.org/10.1016/j.gendis.2020.01.013
Cai L, Wu H, Li D, Zhou K, Zou F (2015) Type 2 diabetes biomarkers of human gut microbiota selected via iterative sure independent screening method. PLoS ONE. https://doi.org/10.1371/journal.pone.0140827
Malik S, Khadgawat R, Anand S et al (2016) Non-invasive detection of fasting blood glucose level via electrochemical measurement of saliva. Springerplus 5:701. https://doi.org/10.1186/s40064-016-2339-6
Nilamyani N, Lawi A, Thamrin SA (2018) A preliminary study on identifying probable biomarker of type 2 diabetes using recursive feature extraction. In: 2018 2nd East Indonesia conference on computer and information technology (EIConCIT), pp 267–270. https://doi.org/10.1109/EIConCIT.2018.8878565
Liu ZY, Ding XP, Bian HJ (2008) Comparisons of properties of tandem repeats associated with beteen diabetes genes and non-diabetes disease genes. In: 2nd international conference on bioinformatics and biomedical engineering, iCBBE 2008, pp 436–440. https://doi.org/10.1109/ICBBE.2008.107
Reddy SS, Sethi N, Rajender R, Mahesh G (2020) Extensive analysis of machine learning algorithms to early detection of diabetic retinopathy. Mater Today Proc. https://doi.org/10.1016/j.matpr.2020.10.894
Kavakiotis I, Tsave O, Salifoglou A, Maglaveras N, Vlahavas I, Chouvarda I (2017) Machine learning and data mining methods in diabetes research. Comput Struct Biotechnol J 15:104–116. https://doi.org/10.1016/j.csbj.2016.12.005
Sikder N, Masud M, Bairagi AK, Arif ASM, Nahid A-A, Alhumyani HA (2021) Severity classification of diabetic retinopathy using an ensemble learning algorithm through analyzing retinal images. Symmetry 13:670
Islam MT, Raihan M, Aktar N, Alam MS, Ema RR, Islam T (2020) Diabetes mellitus prediction using different ensemble machine learning approaches. In: 2020 11th international conference on computing, communication and networking technologies (ICCCNT), pp 1–7
Islam MT, Raihan M, Farzana F, Aktar N, Ghosh P, Kabiraj S (2020) Typical and non-typical diabetes disease prediction using random forest algorithm. In: 2020 11th International conference on computing, communication and networking technologies (ICCCNT), pp 1–6
“Ensembl Genbank”. Available: https://www.ensembl.org/index.html. Accessed 04 Apr 2020
Das B, Turkoglu I (2018) A novel numerical mapping method based on entropy for digitizing DNA sequences. Neural Comput Appl 29:207–215. https://doi.org/10.1007/s00521-017-2871-5
Daş B (2018) Development of new approaches based on signal processing for disease diagnosis from Dna sequences, Fırat University, PhD Thesis, 2018
Grandhi DG, Kumar CV (2007) 2-Simplex mapping for identifying the protein coding regions in DNA. In: TENCON 2007- 2007 IEEE reg. 10 conf., pp 1–3. IEEE
Chakraborty S, Gupta V (2016) DWT Based cancer identification using EIIP. In: 2016 second international conference on computational intelligence communication technology (CICT), pp 718–723. https://doi.org/10.1109/CICT.2016.148
Akhtar M, Epps J, Ambikairajah E (2007) On DNA numerical representations for period-3 based exon prediction. In: 2007 IEEE international workshop on genomic signal processing and statistics, pp 1–4. IEEE
Cristea PD (2002) Conversion of nucleotides sequences into genomic signals. J Cell Mol Med 6:279–303. https://doi.org/10.1111/j.1582-4934.2002.tb00196.x
Cristea PD (2005) Representation and Analysis of DNA sequences. Genomic signal processing and statistics. Eurasip B Ser Signal Process Commun 15–66
Yosinski J, Clune Y, Lipson BH (2014) How transferable are features in deep neural networks?. Adv Neural Inf Process Syst. http://arxiv.org/abs/1411.1792
Ozcan T, Basturk A (2019) Transfer learning-based convolutional neural networks with heuristic optimization for hand gesture recognition. Neural Comput Appl 31:8955–8970. https://doi.org/10.1007/s00521-019-04427-y
Zeiler MD, Fergus R (2014) Visualizing and understanding convolutional networks. In: Lect. Notes Comput. Sci. (Including Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinformatics), pp 818–833
Ullah I, Hussain M, Qazi E-H, Aboalsamh H (2018) An automated system for epilepsy detection using EEG brain signals based on deep learning approach. Expert Syst Appl 107:61–71. https://doi.org/10.1016/j.eswa.2018.04.021
Gopalakrishnan K, Khaitan SK, Choudhary A, Agrawal A (2017) Deep Convolutional Neural Networks with transfer learning for computer vision-based data-driven pavement distress detection. Constr Build Mater 157:322–330. https://doi.org/10.1016/j.conbuildmat.2017.09.110
Simonyan K, Zisserman A (2015) Very deep convolutional networks for large-scale image recognition. arXiv:1409.1556 [cs]
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE computer society conference on computer vision and pattern recognition
Reddy N, Rattani A, Derakhshani R (2018) Comparison of deep learning models for biometric-based mobile user authentication. In: 2018 IEEE 9th international conference on biometrics theory, applications and systems (BTAS), pp 1–6. https://doi.org/10.1109/BTAS.2018.8698586
Chen Z, Cen J, Xiong J (2020) Rolling bearing fault diagnosis using time-frequency analysis and deep transfer convolutional neural network. IEEE Access 8:150248–150261. https://doi.org/10.1109/ACCESS.2020.3016888
Dilmen E, Beyhan S (2017) A novel online LS-SVM approach for regression and classification. IFAC-PapersOnLine 50(1):8642–8647. https://doi.org/10.1016/j.ifacol.2017.08.1521
Khairandish MO, Sharma M, Jain V, Chatterjee JM, Jhanjhi NZ (2021) A Hybrid CNN-SVM threshold segmentation approach for tumor detection and classification of MRI brain images. IRBM. https://doi.org/10.1016/j.irbm.2021.06.003
Baby Saral G, Priya R (2021) Digital screen addiction with KNN and -Logistic regression classification. Mater Today Proc. https://doi.org/10.1016/j.matpr.2020.11.360
Wang Y, Pan Z, Dong J A new two-layer nearest neighbor selection method for kNN classifier—ScienceDirect. https://www.sciencedirect.com/science/article/pii/S0950705121008662. Accessed 07 Feb 2022
Funding
There is no funding source for this article.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author declares that there are no known competing financial interests or personal relationships that could appear to influence the work reported in this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Das, B. A deep learning model for identification of diabetes type 2 based on nucleotide signals. Neural Comput & Applic 34, 12587–12599 (2022). https://doi.org/10.1007/s00521-022-07121-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-022-07121-8