Abstract
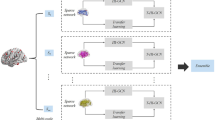
Functional brain connectivity data extracted from functional magnetic resonance imaging (fMRI), characterized by high dimensionality and nonlinear structure, has been widely used to mine the organizational structure for different brain diseases. It is difficult to achieve effective performance by directly using these data for unsupervised clustering analysis of brain diseases. To tackle this problem, in this paper, we propose a dual-stream encoder neural networks with spectral constraint framework for clustering the functional brain connectivity data. Specifically, we consider two different information while encoding the input data: (1) the information between the neighboring nodes, (2) the discriminative features, then design a spectral constraint module to guide the clustering of embedded nodes. The framework contains four modules, Graph Convolutional Encoder, Hard Assignment Optimization Network, Decoder module, and Spectral Constraint module. We train four modules jointly and implement a deep clustering network framework. We conducted experimental analysis on different public functional brain connectivity datasets for evaluating the proposed deep learning clustering model. Compared with the existing unsupervised clustering analysis methods for the brain connectivity data and related deep learning clustering methods, experiments on seven real brain connectivity datasets demonstrate the effectiveness and advantages of our proposed method. The source code is available at https://github.com/hulu88/DENs-SCC.





Similar content being viewed by others
References
Allen EA, Damaraju E, Plis SM, Erhardt EB, Eichele T, Calhoun VD (2014) Tracking whole-brain connectivity dynamics in the resting state. Cereb Cortex 24(3):663–676
Baumgartner R, Scarth G, Teichtmeister C, Somorjai R, Moser E (1997) Fuzzy clustering of gradient-echo functional mri in the human visual cortex. part i: reproducibility. J Magn Reson Imaging 7(6):1094–1101
Biswal B, Zerrin Yetkin F, Haughton VM, Hyde JS (1995) Functional connectivity in the motor cortex of resting human brain using echo-planar mri. Magn Reson Med 34(4):537–541
Bo D, Wang X, Shi C, Zhu M, Lu E, Cui P(2020) Structural deep clustering network. In: Proceedings of The Web Conference pp 1400–1410
Craddock RC, James GA, Holtzheimer PE III, Hu XP, Mayberg HS (2012) A whole brain fmri atlas generated via spatially constrained spectral clustering. Hum Brain Mapp 33(8):1914–1928
Cui G, Zhou J, Yang C, Liu Z Adaptive graph encoder for attributed graph embedding. In: Proceedings of the 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, pp 976–985
Cui X, Xiang J, Guo H, Yin G, Zhang H, Lan F, Chen J (2018) Classification of alzheimer’s disease, mild cognitive impairment, and normal controls with subnetwork selection and graph kernel principal component analysis based on minimum spanning tree brain functional network. Front Comput Neurosci 12:31
Cui X, Xiao J, Guo H, Wang B, Li D, Niu Y, Xiang J, Chen J (2020) Clustering of brain function network based on attribute and structural information and its application in brain diseases. Front Neuroinform 13:79
Jiang Z, Zheng Y, Tan H, Tang B, Zhou H (2016) Variational deep embedding: a generative approach to clustering. CoRR, abs/1611.05148 1
Kazi A, Shekarforoush S, Krishna S.A, Burwinkel H, Vivar G, Kortüm K, Ahmadi S.A, Albarqouni S, Navab N (2019) Inceptiongcn: receptive field aware graph convolutional network for disease prediction. In: International Conference on Information Processing in Medical Imaging, pp 73–85. Springer
Kipf TN, Welling M (2016) Semi-supervised classification with graph convolutional networks. arXiv preprint arXiv:1609.02907
Kipf TN, Welling M (2016) Variational graph auto-encoders. arXiv preprint arXiv:1611.07308
Lanka P, Rangaprakash D, Dretsch MN, Katz JS, Denney TS, Deshpande G (2020) Supervised machine learning for diagnostic classification from large-scale neuroimaging datasets. Brain Imaging Behav 14(6):2378–2416
Lanka P, Rangaprakash D, Gotoor SSR, Dretsch MN, Katz JS, Denney TS Jr, Deshpande G (2020) Malini (machine learning in neuroimaging): a matlab toolbox for aiding clinical diagnostics using resting-state fmri data. Data Brief 29:105213
Li X, Zhang H, Zhang R (2020) Adaptive graph auto-encoder for general data clustering. arXiv preprint arXiv:2002.08648
MacQueen J, et al (1967) Some methods for classification and analysis of multivariate observations. In: Proceedings of the fifth Berkeley symposium on mathematical statistics and probability, vol. 1, pp 281–297. Oakland, CA, USA
Makhzani A, Shlens J, Jaitly N, Goodfellow I, Frey B (2015) Adversarial autoencoders. arXiv preprint arXiv:1511.05644
Moretti P, Muñoz MA (2013) Griffiths phases and the stretching of criticality in brain networks. Nat Commun 4(1):1–10
Ng A, Jordan M, Weiss Y (2001) On spectral clustering: analysis and an algorithm. Adv Neural Inf Process Syst 14:849–856
Ota K, Oishi N, Ito K, Fukuyama H, Group SJS et al (2014) A comparison of three brain atlases for mci prediction. J Neurosci Methods 221:139–150
Pan S, Hu R, Long G, Jiang J, Yao L, Zhang C (2018) Adversarially regularized graph autoencoder for graph embedding. arXiv preprint arXiv:1802.04407
Parisot S, Ktena SI, Ferrante E, Lee M, Guerrero R, Glocker B, Rueckert D (2018) Disease prediction using graph convolutional networks: application to autism spectrum disorder and alzheimer’s disease. Med Image Anal 48:117–130
Snyder LH, Batista A, Andersen RA (1997) Coding of intention in the posterior parietal cortex. Nature 386(6621):167–170
Song C, Liu F, Huang Y, Wang L, Tan T (2013) Auto-encoder based data clustering. In: Iberoamerican congress on pattern recognition, pp 117–124. Springer
Tian F, Gao B, Cui Q, Chen E, Liu TY (2014) Learning deep representations for graph clustering. In: Proceedings of the AAAI Conference on Artificial Intelligence, vol. 28
Wang C, Pan S, Hu R, Long G, Jiang J, Zhang C (2019) Attributed graph clustering: a deep attentional embedding approach. arXiv preprint arXiv:1906.06532
Wang S, He L, Cao B, Lu C.T, Yu PS, Ragin AB (2017) Structural deep brain network mining. In: Proceedings of the 23rd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, pp 475–484
Xie J, Girshick R, Farhadi A Unsupervised deep embedding for clustering analysis. In: International conference on machine learning, pp 478–487
Yang X, Deng C, Zheng F, Yan J, Liu W Deep spectral clustering using dual autoencoder network. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp 4066–4075
Zhao X, Rangaprakash D, Denney TS Jr, Katz JS, Dretsch MN, Deshpande G (2019) Identifying neuropsychiatric disorders using unsupervised clustering methods: data and code. Data Brief 22:570–573
Zhao X, Rangaprakash D, Yuan B, Denney TS Jr, Katz JS, Dretsch MN, Deshpande G (2018) Investigating the correspondence of clinical diagnostic grouping with underlying neurobiological and phenotypic clusters using unsupervised machine learning. Front Appl Math Stat 4:25
Zhuang C, Ma Q (2018) Dual graph convolutional networks for graph-based semi-supervised classification. In: Proceedings of the 2018 World Wide Web Conference, pp 499–508
Acknowledgements
This work is in part supported by the National Science Foundation of China (No. 62006098) and the Postgraduate Research & Practical Innovation Program of Jiangsu Province (SJCX21_1696).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lu, H., Jin, T. Dual-stream encoder neural networks with spectral constraint for clustering functional brain connectivity data. Neural Comput & Applic 34, 12737–12747 (2022). https://doi.org/10.1007/s00521-022-07122-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-022-07122-7