Abstract
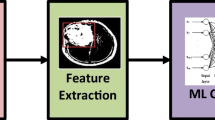
Gastric cancer is the sixth most common cancer and the fourth leading cause of cancer deaths worldwide. Gastric cancer presents with a more insidious onset and is most frequently discovered at an advanced stage. Early diagnosis is critical since the stage of the disease is determinant in the severity, treatment, and survival rate of cancer. In the study, the Region of Interest (RoI) was determined in histopathological images using image preprocessing techniques and signet ring cell carcinoma (SRCC) was detected with popular deep learning models VGG16, VGG19, and InceptionV3. The fine-tuning strategy was applied by customizing the last five layers of deep network models based on the target data. The parameters of accuracy, precision, recall, and F1-score were used to evaluate the model performance. Signet ring cell dataset taken from the competition “Digestive System Pathological Detection, and Segmentation Challenge 2019” was employed. When compared to results of the DigestPath2019 Grand challenge ring cell gastric cancer competition, higher accuracy rates were obtained using deep learning models with the accurate defined RoI images. VGG16 model exhibited a higher performance with accuracy of 95% and a F1-score of 95% among the models. The results obtained by the algorithm were analyzed and confirmed by the experienced pathologist.










Similar content being viewed by others
References
https://www.who.int/news-room/fact-sheets/detail/cancer, Word Health Organization
Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J (2016) Cancer statistics in China, 2015. CA Cancer J Clin 66(2):115–132
Henson DE, Dittus C, Younes M, Nguyen H, Albores-Saavedra J (2004) Differential trends in the intestinal and diffuse types of gastric carcinoma in the United States, 1973–2000: increase in the signet ring cell type. Arch Pathol Lab Med 128(7):765–770
Lauwers G, Carneiro F, Graham D, Curado M, Franceschi S (2010) Classification of tumours of the digestive system. IARC Press, Lyon, pp 48–58
Gao JM, Tang SS, Fu W, Fan R (2009) Signet-ring cell carcinoma of ampulla of Vater: contrast-enhanced ultrasound findings. World J Gastroenterol: WJG 15(7):888
Association Japanese Gastric Cancer (1999) Japanese classification of gastric carcinoma: Kanehara. Tokyo, Japan
El-Zimaity HM, Itani K, Graham DY (1997) Early diagnosis of signet ring cell carcinoma of the stomach: role of the Genta stain. J Clin Pathol 50(10):867–868
Hosokawa O, Hattori M, Douden K, Hayashi H, Ohta K, Kaizaki Y (2007) Difference in accuracy between gastroscopy and colonoscopy for detection of cancer. Hepatogastroenterology 54(74):442–444
Sano T, Coit DG, Kim HH, Roviello F, Kassab P, Wittekind C, Yamamoto Y, Ohashi Y (2017) Proposal of a new stage grouping of gastric cancer for TNM classification: international gastric cancer association staging project. Gastric Cancer 20(2):217–225
Wang J, Yu JC, Kang WM, Ma ZQ (2012) Treatment strategy for early gastric cancer. Surg Oncol 21(2):119–123
Tsubono Y, Hisamichi S (2000) Screening for gastric cancer in Japan. Gastric Cancer 3(1):9–18
Fass L (2008) Imaging and cancer: a review. Mol Oncol 2(2):115–152
Doi K (2007) Computer-aided diagnosis in medical imaging: historical review, current status and future potential. Comput Med Imaging Graph 31(4–5):198–211
Schmidt-Erfurth U, Sadeghipour A, Gerendas BS, Waldstein SM, Bogunović H (2018) Artificial intelligence in retina. Prog Retin Eye Res 67:1–29
Hirasawa T, Aoyama K, Tanimoto T, Ishihara S, Shichijo S, Ozawa T, Ohnishi T, Fujishiro M, Matsua K, Fujisaki J, Tada T (2018) Application of artificial intelligence using a convolutional neural network for detecting gastric cancer in endoscopic images. Gastric Cancer 21(4):653–660
Budak C, Mençik V, Gider V (2021) Determining similarities of COVID-19–lung cancer drugs and affinity binding mode analysis by graph neural network-based GEFA method. J Biomol Struct Dyn. https://doi.org/10.1080/07391102.2021.2010601
Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, Webster DR (2016) Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 316(22):2402–2410
Byrne MF, Chapados N, Soudan F, Oertel C, Pérez ML, Kelly R, Igbal N, Chandelier F, Rex DK (2019) Real-time differentiation of adenomatous and hyperplastic diminutive colorectal polyps during analysis of unaltered videos of standard colonoscopy using a deep learning model. Gut 68(1):94–100
Vives-Boix V, Ruiz-Fernández D (2021) Fundamentals of artificial metaplasticity in radial basis function networks for breast cancer classification. Neural Comput Appl 33:12869–12880
Malon CD, Cosatto E (2013) Classification of mitotic figures with convolutional neural networks and seeded blob features. J Pathol Inform 4:1–9
Moazzen Y, Capar A, Albayrak A, Çalık N, Töreyin BU (2019) Metaphase finding with deep convolutional neural networks. Biomed Signal Process Control 52:353–361
Hu Q, Qin A, Zhang Q, He J, Sun G (2018) Fault diagnosis based on weighted extreme learning machine with wavelet packet decomposition and KPCA. IEEE Sens J 18(20):8472–8483
Xiao C, Yu M, Zhang B, Wang H, Jiang C (2020) Discrete component prognosis for hybrid systems under intermittent faults. IEEE Trans Autom Sci Eng 18(4):1766–77
Budak C, Türk M, Toprak A (2015) Reduction in impulse noise in digital images through a new adaptive artificial neural network model. Neural Comput Appl 26(4):835–843
Albayrak A, Bilgin G (2019) Automatic cell segmentation in histopathological images via two-staged superpixel-based algorithms. Med Biol Eng Compu 57(3):653–665
Spanhol FA, Oliveira LS, Cavalin PR, Petitjean C, Heutte L (2017) Deep features for breast cancer histopathological image classification. In 2017 IEEE International conference on systems, man, and cybernetics (SMC), (pp. 1868–1873)
Bardou D, Zhang K, Ahmad SM (2018) Classification of breast cancer based on histology images using convolutional neural networks. Ieee Access 6:24680–24693
Kanavati F, Ichihara S, Rambeau M, Iizuka O, Arihiro K, Tsuneki M (2020) Deep learning models for gastric signet ring cell carcinoma classification in whole slide images. arXiv preprint arXiv:2011.09247
Harsono IW, Liawatimena S, Cenggoro TW (2020) Lung nodule detection and classification from thorax ct-scan using retinanet with transfer learning. J King Saud Univ-Comput Infor Sci
Pham HHN, Futakuchi M, Bychkov A, Furukawa T, Kuroda K, Fukuoka J (2019) Detection of lung cancer lymph node metastases from Whole-Slide histopathologic images using a two-step deep learning approach. Am J Pathol 189(12):2428–2439
Xu Y, Jia Z, Ai Y, Zhang F, Lai M, Eric I, Chang C (2015) Deep convolutional activation features for large scale brain tumor histopathology image classification and segmentation. In 2015 IEEE international conference on acoustics, speech and signal processing (ICASSP), (pp. 947–951)
Chang YH, Thibault G, Madin O, Azimi V, Meyers C, Johnson B, Gray JW (2017) Deep learning based Nucleus Classification in pancreas histological images. In 2017 39th annual international conference of the IEEE engineering in medicine and biology society (EMBC), (pp. 672–675)
Tambe R, Mahajan S, Shah U, Agrawal M, Garware B (2019). Towards designing an automated classification of lymphoma subtypes using deep neural networks. In Proceedings of the ACM India joint international conference on data science and management of data, (pp. 143–149)
Wang D, Gu C, Wu K, Guan X (2017) Adversarial neural networks for basal membrane segmentation of microinvasive cervix carcinoma in histopathology images. In 2017 International conference on machine learning and cybernetics (ICMLC), 2, (pp. 385–389)
Ponzio F, Macii E, Ficarra E, Di Cataldo S (2018) Colorectal cancer classification using deep convolutional networks. In Proceedings of the 11th international joint conference on biomedical engineering systems and Technologies, 2, (pp. 58–66)
https://digestpath2019.grandchallenge.org/, (2019) Digestive-system pathological detection and segmentation challenge
Achanta R, Shaji A, Smith K, Lucchi A, Fua P, Süsstrunk S (2012) SLIC superpixels compared to state-of-the-art superpixel methods. IEEE Trans Pattern Anal Mach Intell 34(11):2274–2282
Simonyan K, Zisserman (2014) Very deep convolutional networks for large-scale image recognition. arXiv, 1409
Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z (2016) Rethinking the inception architecture for computer vision. In Proceedings of the IEEE conference on computer vision and pattern recognition, (pp 2818–2826)
Macenko M, Niethammer M, Marron JS, Borland D, Woosley JT, Guan X, Thomas, NE (2009) A method for normalizing histology slides for quantitative analysis. In 2009 IEEE International symposium on biomedical imaging: From Nano to Macro, (pp. 1107–1110)
Ciregan D, Meier U, Schmidhuber J (2012) Multi-column deep neural networks for image classification. In 2012 IEEE conference on computer vision and pattern recognition, (pp. 3642–3649)
Li Q, Cai W, Wang X, Zhou Y, Feng DD, Chen M (2014) Medical image classification with convolutional neural network. In 2014 13th international conference on control automation robotics & vision (ICARCV), (pp. 844–848)
Krizhevsky A, Sutskever I, Hinton GE (2017) ImageNet classification with deep convolutional neural networks. Commun ACM 60(6):84–90
Ioffe S, Szegedy C (2015) Batch normalization: accelerating deep network training by reducing internal covariate shift. In International conference on machine learning, (pp. 448–456)
Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R (2014) Dropout: a simple way to prevent neural networks from overfitting. J Machine Learn Res 15(1):1929–1958
Fang Z, Zhang W, Ma H (2019). Breast cancer classification with ultrasound images based on SLIC. arXiv preprint arXiv:1904.11322.
Dhanachandra N, Manglem K, Chanu YJ (2015) Image segmentation using K-means clustering algorithm and subtractive clustering algorithm. Proc Comput Sci 54:764–771
Win KY, Choomchuay S, Hamamoto K, Raveesunthornkiat M, Rangsirattanakul L, Pongsawat S (2018) Computer aided diagnosis system for detection of cancer cells on cytological pleural effusion images. BioMed Res Int 2018:1–21
Ying H, Song Q, Chen J, Liang T, Gu J, Zhuang F, Wu J (2021) A semi-supervised deep convolutional framework for signet ring cell detection. Neurocomputing 453:347–356
Lin T, Guo Y, Yang C, Yang J, Xu Y (2021) Decoupled gradient harmonized detector for partial annotation: application to signet ring cell detection. Neurocomputing. 453:337–346
Li J, Yang S, Huang X, Da Q, Yang X, Hu Z, Li H (2019) Signet ring cell detection with a semi-supervised learning framework. In International conference on information processing in medical imaging, (pp. 842–854)
Zhang S, Yuan Z, Wang Y, Bai Y, Chen B, Wang H (2021) REUR: a unified deep framework for signet ring cell detection in low-resolution pathological images. Comput Biol Med 136:104711
Wang S, Jia C, Chen Z, Gao X (2020) Signet ring cell detection with classification reinforcement detection network. In International symposium on bioinformatics research and applications, (pp. 13–25)
Acknowledgements
We are grateful to Assoc. Prof. Dr. Ulaş ALABALIK for his support in confirming and interpreting the results obtained in this study (staff at Department of Medical Pathology, Faculty of Medicine, Dicle University)
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Budak, C., Mençik, V. Detection of ring cell cancer in histopathological images with region of interest determined by SLIC superpixels method. Neural Comput & Applic 34, 13499–13512 (2022). https://doi.org/10.1007/s00521-022-07183-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-022-07183-8