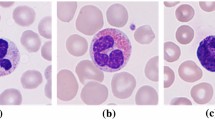
Abstract
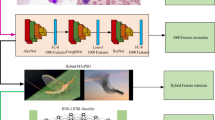
Blood cell count is an important parameter in analysing a person’s health condition. White blood corpuscles (leukocytes) are responsible for deciding the immunity system of a person. White blood cells (WBC) are classified into: lymphocytes, monocytes and granulocytes. Granulocytes are sub-classified into: neutrophils, eosinophils, and basophils. These five types perform different functions in acting as a defence mechanism of the body. Manual investigation performed at laboratories for WBC count is prone to errors due to certain factors like human fatigue, inter-operability errors, etc. Further, there is serious issue in using trained data which cater to the changes in morphology of the white blood corpuscles, in order that trained classifiers could capitalize well. In a way to reduce misclassification rate, a methodology wherein a deep learning approach integrated with an evolutionary algorithm is proposed. Convolutional neural network (CNN) hyper-parameters were optimized using particle swarm optimization algorithm (PSO) to improve the network performance in classifying white blood cells into five types. The method is tested on merged LISC and BCCD datasets which achieved classification accuracy of 99.2% with 94.56% sensitivity, 98.78% specificity and 0.982 AUC. The results are compared with similar proven algorithms like genetic algorithms (GA), differential evolution (DE) and grey wolf optimization (GWO) algorithms. The experimental outcomes demonstrated PSO’s potential in optimizing the CNN hyper-parameters for white blood cell classification enhancing the sensitivity rate and serve a best second opinion in assessing blood cell count.





Similar content being viewed by others
References
Liu S, Deng Z, Li J, Wang J, Huang N, Cui R, Zhang Q, Mei J, Zhou W, Zhang C, Ye Q, Tian J (2019) Measurement of the refractive index of whole blood and its components for a continuous spectral region. J Biomed Opt 24(3):1–5. https://doi.org/10.1117/1.JBO.24.3.035003
https://www.britannica.com/science/blood-biochemistry/Plasma#ref257801. Accessed 13 April 2021
Herron, Connie MS, RN-BC, NP-C, MDI. (2012) Know your WBCs, Nursing Made Incredibly Easy. 10(1):11–15. https://doi.org/10.1097/01.NME.0000408238.43869.e2
https://www.medicalnewstoday.com/articles/327446 (accessed: 13th April 2021)
https://www.open.edu/openlearn/ocw/mod/oucontent/view.php?id=65372§ion=3.1. Accessed 13 April 2021
Shafique S, Tehsin S (2018) Computer-aided diagnosis of acute lymphoblastic leukaemia. Comput Math Methods Med 2018:6125289. https://doi.org/10.1155/2018/6125289
Habibzadeh M, Krzyżak A, Fevens T (2013) Comparative study of shape, intensity and texture features and support vector machine for white blood cell classification. Appl Comput Sci 7:20–35
Duan Y, Wang J, Hu M, Zhou M, Li Q, Sun L, Qiu S, Wang Y (2019) Leukocyte classification based on spatial and spectral features of microscopic hyperspectral images. Opt Laser Technol
Jiang M, Cheng L, Qin F, Du L, Zhang M (2018) White blood cells classification with deep convolutional neural networks. Int J Pattern Recognit Artif Intell 32: 1857006:1–1857006:19.
Liang G, Hong H, Xie W, Zheng L (2018) Combining convolutional neural network with recursive neural network for blood cell image classification. IEEE Access 6:36188–36197. https://doi.org/10.1109/access.2018.2846685
Hegde RB, Prasad K, Hebbar H, Singh B (2018) Development of a robust algorithm for detection of nuclei and classification of white blood cells in peripheral blood smear images. J Med Syst 42(6):110. https://doi.org/10.1007/s10916-018-0962-1
Sahlol AT, Kollmannsberger P, Ewees AA (2020) Efficient classification of white blood cell leukemia with improved swarm optimization of deep features. Sci Rep 10(1):2536. https://doi.org/10.1038/s41598-020-59215-9
Prinyakupt J, Pluempitiwiriyawej C (2015) Segmentation of white blood cells and comparison of cell morphology by linear and naïve Bayes classifiers. BioMed Eng 14
Tingting H, Jianwei Z, Feilong C (2015) A classification algorithm for white blood cells based on the synthetic feature and random forest. J China Univ Metro 26(4):474–479
Macawile MJ, Quinones VV, Ballado A, Cruz JD, Caya MV (2018) White blood cell classification and counting using convolutional neural network. In: 2018 3rd international conference on control and robotics engineering (ICCRE). doi:https://doi.org/10.1109/iccre.2018.8376476
Zhao J, Zhang M, Zhou Z, Chu J, Cao F (2016) Automatic detection and classification of leukocytes using convolutional neural networks. Med Biol Eng Compu 55:1287–1301
Hegde RB, Prasad K, Hebbar H, Singh BM (2019) Comparison of traditional image processing and deep learning approaches for classification of white blood cells in peripheral blood smear images. Biocybern Biomed Eng 39(12):382–392
Habibzadeh M, Jannesari M, Rezaei Z, Baharvand H, Totonchi M (2018) Automatic white blood cell classification using pre-trained deep learning models: ResNet and Inception. In: International conference on machine vision. Proc SPIE Int Soc Opt Eng 10696:1069612
Kurniadi FI, Putri VK (2019) A comparison of human crafted features and machine crafted features on white blood cells classification. J Phys Conf Ser 1201(1):012045
Srivastava N, Hinton GE, Krizhevsky A, Sutskever I, Salakhutdinov R (2014) Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res 15:1929–1958
Sun D, Wang M, Li A (2019) A multimodal deep neural network for human breast cancer prognosis prediction by integrating multi-dimensional data. IEEE/ACM Trans Comput Biol Bioinf 16(3):841–850
Acharya UR, Oh SL, Hagiwara Y, Tan JH, Adeli H (2018) Deep convolutional neural network for the automated detection and diagnosis of seizure using EEG signals. Comput Biol Med 100:270–278. https://doi.org/10.1016/j.compbiomed.2017.09.017
Wang G, Li W, Zuluaga MA, Pratt R, Patel PA, Aertsen M, Doel T, David AL, Deprest J, Ourselin S, Vercauteren T (2018) Interactive medical image segmentation using deep learning with image-specific fine tuning. IEEE Trans Med Imaging 37(7):1562–1573. https://doi.org/10.1109/TMI.2018.2791721
Piuri V, Scotti F (2004) Morphological classification of blood leucocytes by microscope images. In: 2004 IEEE international conference on computational intelligence for measurement systems and applications, 2004. CIMSA, pp 103–108
Hiremath PS, Bannigidad P, Geeta S (2010) Automated identification and classification of white blood cells (Leukocytes) in digital microscopic images. Int J Comput Appl, pp 59–63
Patil A, Patil MD, Birajdar GK (2020) White blood cells image classification using deep learning with canonical correlation analysis. Irbm. https://doi.org/10.1016/j.irbm.2020.08.005
Throngnumchai K, Lomvisai P, Tantasirin C, Phasukkit P (2019) Classification of White blood cell using Deep Convolutional Neural Network. In: 2019 12th biomedical engineering international conference (BMEiCON), pp 1–4
Sengur A, Akbulut Y, Budak U, Comert Z (2019) White blood cell classification based on shape and deep features. In: 2019 international artificial intelligence and data processing symposium (IDAP). doi:https://doi.org/10.1109/idap.2019.8875945
Kutlu H, Avci E, Özyurt F (2020) White blood cells detection and classification based on regional convolutional neural networks. Med Hypotheses 135:109472. https://doi.org/10.1016/j.mehy.2019.109472
Edraki A, Razminia A (2018) Classification of white blood cells using convolutional neural network. Iranian South Med J 21:65–80
Yao X, Sun K, Xixi Bu, Zhao C, Jin Yu (2021) Classification of white blood cells using weighted optimized deformable convolutional neural networks. Artif Cells Nanomed Biotechnol 49(1):147–155. https://doi.org/10.1080/21691401.2021.1879823
Baydilli YY, Atila Ü (2020) Classification of white blood cells using capsule networks. Comput Med Imaging Graph. https://doi.org/10.1016/j.compmedimag.2020.101699
Toğaçar M, Ergen B, Cömert Z (2020) Classification of white blood cells using deep features obtained from convolutional neural network models based on the combination of feature selection methods. Appl Soft Comput 97:106810
Özyurt F (2020) A fused CNN model for WBC detection with MRMR feature selection and extreme learning machine. Soft Comput 24:8163–8172
Çınar A, Tuncer SA (2021) Classification of lymphocytes, monocytes, eosinophils, and neutrophils on white blood cells using hybrid Alexnet-GoogleNet-SVM. SN Appl Sci 3:503. https://doi.org/10.1007/s42452-021-04485-9
Malkawi A, Al-Assi A, Salameh R, Alquran T, Alqudah H (2020) White blood cells classification using convolutional neural network hybrid system. In 2020 IEEE 5th middle east and Africa conference on biomedical engineering (MECBME), pp 1–5
Deepak G, Jatin A, Utkarsh A, Khanna A, Albuquerque VHC (2019) Optimized binary bat algorithm for classification of white blood cells. Measurement, 143
Semerjian S, Khong YF, Mirzaei S (2021) White blood cells classification using built-in customizable trained convolutional neural network. In: Proceedings of the 2021 international conference on emerging smart computing and informatics (ESCI), Pune, India, 5–7 March 2021, pp 357–362
Ridoy MA, Islam MR (2020) An automated approach to white blood cell classification using a lightweight convolutional neural network. In: 2020 2nd international conference on advanced information and communication technology (ICAICT), pp 480–483
The LISC dataset (2019). LISC: leukocyte images for segmentation and classification, http://users.cecs.anu.edu.au/∼hrezatofighi/Data/Leukocyte%20Data.html
Rezatofighi SH, Soltanian-Zadeh H (2011) Automatic recognition of five types of white blood cells in peripheral blood. Comput Med Imaging Graph 35(4):333–343. https://doi.org/10.1016/j.compmedimag.2011.01.003
BCCD, Blood Cell Images. Available online: URL https://www.kaggle.com/paultimothymooney/bloodcells/home
Shelhamer E, Long J, Darrell T (2017) Fully convolutional networks for semantic segmentation. IEEE Trans Pattern Anal Mach Intell 39(4):640–651. https://doi.org/10.1109/TPAMI.2016.2572683
Johnson JM, Khoshgoftaar TM (2019) Survey on deep learning with class imbalance. J Big Data 6:1–54
LeCun Y, Bengio Y (1998) Convolutional networks for images, speech, and time series. The handbook of brain theory and neural networks. MIT Press, Cambridge, MA, pp 255–258
LeCun Y, Kavukcuoglu K, Farabet C (2010) Convolutional networks and applications in vision. In: Proceedings of 2010 IEEE international symposium on circuits and systems, pp 253–256.
Hijazi SL, Kumar R, Rowen C (2015) Using convolutional neural networks for image recognition. Technical Report
Eberhart RC, Kennedy J (1995) A new optimizer using particle swarm theory. MHS'95. In: Proceedings of the sixth international symposium on micro machine and human science, pp 39–43
Eberhart RC, Shi Y (2001) Particle swarm optimization: developments, applications and resources. In: Proceedings of the 2001 congress on evolutionary computation (IEEE Cat. No.01TH8546), vol 1, pp 81–86
Eberhart RC, Shi Y (2000) Comparing inertia weights and constriction factors in particle swarm optimization. In: Proceedings of the 2000 congress on evolutionary computation. CEC00 (Cat. No.00TH8512), vol 1, pp 84–88
Banik PP, Saha R, Kim K (2020) An automatic nucleus segmentation and CNN model based classification method of white blood cell. Expert Syst Appl 149:113211
Cai X, Cui Z, Zeng J, Tan Y (2009) Individual parameter selection strategy for particle swarm optimization, particle swarm optimization, Aleksandar Lazinica, IntechOpen, Doi: https://doi.org/10.5772/6742. Available from: https://www.intechopen.com/chapters/6255
Shi Y, Eberhart R (1998) Parameter selection in particle swarm optimization. In Evolutionary programming VIZ: Proceedings of EP98. Springer, New York, pp 591–600
Rubinstein RY, Kroese DP (2004) The cross entropy method: a unified approach to combinatorial optimization, monte-carlo simulation and machine learning (information science and statistics). Springer, Berlin
Glorot X, Bengio Y (2010) Understanding the difficulty of training deep feedforward neural networks. In: Proceedings of the thirteenth international conference on artificial intelligence and statistics, PMLR vol 9, pp 249–256
LeCun Y, Boser BE, Denker JS, Henderson D, Howard RE, Hubbard WE, Jackel LD (1989) Handwritten digit recognition with a back-propagation network. NIPS. In: Touretzky D (ed) Advances in neural information processing systems (NIPS 1989), Denver, CO. vol 2. Morgan Kaufmann. 1990
Kohavi R (1995) A study of cross-validation and bootstrap for accuracy estimation and model selection. In: Proceedings of the 14th International Joint Conference on Artificial intelligence (IJCAI’95), Canada, vol 2, pp 1137–1143
Balasubramanian K, Ananthamoorthy NP (2020) Improved adaptive neuro-fuzzy inference system based on modified glowworm swarm and differential evolution optimization algorithm for medical diagnosis. Neural Comput Appl. https://doi.org/10.1007/s00521-020-05507-0
Jung C, Abuhamad M, Alikhanov J, Mohaisen A, Han K, Nyang D (2019) W-Net: a CNN-based architecture for white blood cells image classification. ArXiv:abs/1910.01091.
Ghosh A, Singh S, Sheet D (2017) Simultaneous localization and classification of acute lymphoblastic leukemic cells in peripheral blood smears using a deep convolutional network with average pooling layer. In: 2017 IEEE international conference on industrial and information systems (ICIIS), pp 1–6
Habibzadeh M, Jannesari M, Rezaei Z, Baharvand H, Totonchi M (2018) Automatic white blood cell classification using pre-trained deep learning models: ResNet and Inception. In: International conference on machine vision
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors declare no conflict of interest and potential funding.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Balasubramanian, K., Ananthamoorthy, N.P. & Ramya, K. An approach to classify white blood cells using convolutional neural network optimized by particle swarm optimization algorithm. Neural Comput & Applic 34, 16089–16101 (2022). https://doi.org/10.1007/s00521-022-07279-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-022-07279-1