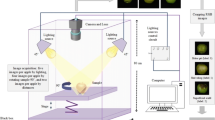
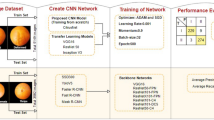
Abstract
Physiological disorders in apples are due to post-harvest conditions. For this reason, automatic identification of physiological disorders is important in obtaining agricultural information. Image processing is one of the techniques that can help achieve the features of physiological disorders. Physiological disorders during image acquisition can be affected by the changes in brightness values created by different lighting conditions. This changes the results of the classification. In recent years, the convolutional neural network (CNN) has been a successful approach in automatically obtaining deep features from raw images in image classification problems. The study aims to classify physiological disorders using machine learning (ML) methods according to extracted deep features of the images under different lighting conditions. The data sets were created by acquired images (1080 images) and augmentation images (4320 images). Deep features were extracted using five popular pre-trained CNN models in these data sets, and these features were classified using five ML methods. The highest average accuracy was obtained with the VGG19(fc6) + SVM method in the data set-1 and data set-2 and were 96.11 and 96.09%, respectively. With this study, physiological disorders can be determined early, and needed precautions can be taken before and after harvest, not too late.







Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.References
Buyukarikan U (2019) Agricultural practices of apple and apple nursery production according to Turkish IAS 41 accounting standard in an agricultural enterprise. Custos E Agronegocio Online 15(2):465–488
Şaşmaz MÜ, Özel Ö (2019) Tarım sektörüne sağlanan mali teşviklerin tarım sektörü gelişimi üzerindeki etkisi: Türkiye örneği. Dumlupınar Üniversitesi Sosyal Bilimler Dergisi 61:50–65
Singh Z, Zaharah SS (2011) Controlled atmosphere storage of mango fruit: challenges and thrusts and its implications in international mango trade. In: Global conference on augmenting production and Utilization of Mango: biotic and abiotic stresses 1066, 179–191. https://doi.org/10.17660/ActaHortic.2015.1066.21
Seppä L, Peltoniemi A, Tahvonen R, Tuorila H (2013) Flavour and texture changes in apple cultivars during storage. LWT-Food Sci Technol 54(2):500–512. https://doi.org/10.1016/j.lwt.2013.06.012
Toivonen PM (2004) Postharvest storage procedures and oxidative stress. HortScience 39(5):938–942
Watkins CB (2017) Postharvest physiological disorders of fresh crops. In: Thomas B, Murray BG, Murphy DJ (eds) Encyclopedia of applied plant sciences. Oxford, Elsevier, pp 315–322
Elgar HJ, Burmeister DM, Watkins CB (1998) Storage and handling effects on a CO2-related internal browning disorder of Braeburn' Apples. HortScience 33(4): 719–722. https://doi.org/10.21273/HORTSCI.33.4.719
Clark CJ, Burmeister DM (1999) Magnetic resonance imaging of browning development in Braeburn' Apple during controlled-atmosphere storage under high CO2. HortScience 34(5):915–919. https://doi.org/10.21273/HORTSCI.34.5.915
Hatoum D, Hertog ML, Geeraerd AH, Nicolai BM (2016) Effect of browning related pre-and postharvest factors on the ‘Braeburn’ apple metabolome during CA storage. Postharvest Biol Technol 111:106–116. https://doi.org/10.1016/j.postharvbio.2015.08.004
Karaçalı İ (2006) Bahçe ürünlerinin muhafaza ve pazarlanması. İzmir, Turkey, Ege Üniversitesi
Swezey SL (2000) Organic apple production manual (vol 3403), UCANR Publications
Özgönen H, Kılıç HÇ (2009) Isparta ili’nde elmalarda sorun olan hasat sonrası hastalıkların ve yaygınlık oranlarının belirlenmesi. Int J Agric Nat Sci 2(2):53–60
Lurie S, Watkins CB (2012) Superficial scald, its etiology and control. Postharvest Biol Technol 65:44–60. https://doi.org/10.1016/j.postharvbio.2011.11.001
Osinenko P, Biegert K, McCormick RJ, Göhrt T, Devadze G, Streif J, Streif S (2021) Application of non-destructive sensors and big data analysis to predict physiological storage disorders and fruit firmness in ‘Braeburn’ apples. Comput Electron Agric 183:106015. https://doi.org/10.1016/j.compag.2021.106015
Şen F, İslam A, Koçak S, Karaçalı İ (2009) Elmada Fizyolojik Bozukluklar. Int J Agric Nat Sci 2(1):121–126
Toivonen PMA, Hodges DM (2011) Abiotic stress in harvested fruits and vegetables. In: Shanker A (ed) Abiotic stress in plants—mechanisms and adaptations. China, InTech
Lu Y, Lu R (2017) Non-destructive defect detection of apples by spectroscopic and imaging technologies: a review. Trans ASABE 60(5):1765–1790. https://doi.org/10.13031/trans.12431
Cunha JB (2003) Application of image processing techniques in the characterization of plant leafs. In: 2003 IEEE international symposium on industrial electronics (Cat No 03TH8692), Rio de Janeiro, Brazil, 612–616. https://doi.org/10.1109/ISIE.2003.1267322
Costa C, Antonucci F, Pallottino F, Aguzzi J, Sun D-W, Menesatti P (2011) Shape analysis of agricultural products: a review of recent research advances and potential application to computer vision. Food Bioprocess Technol 4(5):673–692. https://doi.org/10.1007/s11947-011-0556-0
Türkoğlu M, Hanbay D (2018) Apricot disease identification based on attributes obtained from deep learning algorithms. In: 2018 International Conference on Artificial Intelligence and Data Processing (IDAP) Malatya, Turkey, pp 1–4. https://doi.org/10.1109/IDAP.2018.8620831
Arı B, Arı A, Şengür A, Tuncer SA (2019) Classification of apricot leaves with extreme learning machines using deep features. In: 2019 1st International informatics and software engineering conference (UBMYK), Ankara, Turkey, 1–5. https://doi.org/10.1109/UBMYK48245.2019.8965491
Kafle GK, Khot LR, Jarolmasjed S, Yongsheng S, Lewis K (2016) Robustness of near infrared spectroscopy based spectral features for non-destructive bitter pit detection in honeycrisp apples. Postharvest Biol Technol 120:188–192. https://doi.org/10.1016/j.postharvbio.2016.06.013
Ariana D, Guyer DE, Shrestha B (2006) Integrating multispectral reflectance and fluorescence imaging for defect detection on apples. Comput Electron Agric 50(2):148–161. https://doi.org/10.1016/j.compag.2005.10.002
Lötze E, Huybrechts C, Sadie A, Theron KI, Valcke RM, (2006) Fluorescence imaging as a non-destructive method for pre-harvest detection of bitter pit in apple fruit (Malus domestica Borkh.). Postharvest Biol Technol 40(3):287–294. https://doi.org/10.1016/j.postharvbio.2006.02.004
Jarolmasjed S, Espinoza CZ, Sankaran S (2017) Near infrared spectroscopy to predict bitter pit development in different varieties of apples. J Food Measurement Charact 11(3):987–993. https://doi.org/10.1007/s11694-017-9473-x
Yan Q, Yang B, Wang W, Wang B, Chen P, Zhang J (2020) Apple leaf diseases recognition based on an improved convolutional neural network. Sensors 20(12):3535. https://doi.org/10.3390/s20123535
Shi Y, Huang W, Zhang S (2017) Apple disease recognition based on two-dimensionality subspace learning. Comput Eng Appl 53(22):180–184. https://doi.org/10.3778/j.issn.1002-8331.1605-0073
Zhao W, Yamada W, Li T, Digman M, Runge T (2021) Augmenting crop detection for precision agriculture with deep visual transfer learning: a case study of bale detection. Remote Sens 13(1):23. https://doi.org/10.3390/rs13010023
Dandıl E, Polattimur R (2020) Dog behavior recognition and tracking based on faster R-CNN. J Faculty Eng Architect Gazi Univ 35(2):819–834 https://doi.org/10.17341/gazimmfd.541677
Kamilaris A, Prenafeta-Boldú FX (2018) Deep learning in agriculture: a survey. Comput Electron Agric 147:70–90. https://doi.org/10.1016/j.compag.2018.02.016
Altuntaş Y, Cömert Z, Kocamaz AF (2019) Identification of haploid and diploid maize seeds using convolutional neural networks and a transfer learning approach. Comput Electron Agric 163:104874. https://doi.org/10.1016/j.compag.2019.104874
Reichstein M, Camps-Valls G, Stevens B, Jung M, Denzler J, Carvalhais N, Prabhat (2019) Deep learning and process understanding for data-driven Earth system science. Nature 566:195–204. https://doi.org/10.1038/s41586-019-0912-1
Wang Z, Li M, Wang H, Jiang H, Yao Y, Zhang H, Xin J (2019) Breast cancer detection using extreme learning machine based on feature fusion with CNN deep features. IEEE Access 7:105146–105158. https://doi.org/10.1109/ACCESS.2019.2892795
Nachtigall LG, Araujo RM, Nachtigall GR (2017) Use of images of leaves and fruits of apple trees for automatic identification of symptoms of diseases and nutritional disorders. Int J Monitor Surveill Technol Res 5(2):1–14. https://doi.org/10.4018/IJMSTR.2017040101
Lu Y, Lu R (2018) Detection of surface and subsurface defects of apples using structured-illumination reflectance imaging with machine learning algorithms. Trans ASABE 61(6):1831–1842 https://doi.org/10.13031/trans.12930
Kumar Y, Dubey AK, Arora RR, Rocha A (2020) Multiclass classification of nutrients deficiency of apple using deep neural network. Neural Comput Appl 1–12. https://doi.org/10.1007/s00521-020-05310-x
Lashgari M, Imanmehr A, Tavakoli H (2020) Fusion of acoustic sensing and deep learning techniques for apple mealiness detection. J Food Sci Technol 57(6):2233–2240. https://doi.org/10.1007/s13197-020-04259-y
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521(7553):436–444. https://doi.org/10.1038/nature14539
Toğaçar M, Ergen B, Cömert Z, Özyurt F (2020) A deep feature learning model for pneumonia detection applying a combination of mRMR feature selection and machine learning models. IRBM 41(4):212–222. https://doi.org/10.1016/j.irbm.2019.10.006
Anubha Pearline S, Sathiesh Kumar V, Harini S (2019) A study on plant recognition using conventional image processing and deep learning approaches. J Intell Fuzzy Syst 36(3):1997–2004. https://doi.org/10.3233/JIFS-169911
Kaya A, Keceli AS, Catal C, Yalic HY, Temucin H, Tekinerdogan B (2019) Analysis of transfer learning for deep neural network based plant classification models. Comput Electron Agric 158:20–29. https://doi.org/10.1016/j.compag.2019.01.041
Turkoglu M, Hanbay D (2019) Plant recognition system based on deep features and color-LBP method. In: 2019 27th Signal Processing and Communications Applications Conference (SIU), Sivas, Turkey, 1–4. https://doi.org/10.1109/SIU.2019.8806592
Türkoğlu M, Hanbay D (2019) Combination of deep features and KNN algorithm for classification of leaf-based plant species. In: 2019 International artificial intelligence and data processing symposium (IDAP), Malatya, Turkey. https://doi.org/10.1109/IDAP.2019.8875911
Suh HK, Ijsselmuiden J, Hofstee JW, van Henten EJ (2018) Transfer learning for the classification of sugar beet and volunteer potato under field conditions. Biosys Eng 174:50–65. https://doi.org/10.1016/j.biosystemseng.2018.06.017
Cıbuk M, Budak U, Guo Y, Ince MC, Sengur A (2019) Efficient deep features selections and classification for flower species recognition. Measurement 137:7–13. https://doi.org/10.1016/j.measurement.2019.01.041
Toğaçar M, Ergen B, Cömert Z (2020) Classification of flower species by using features extracted from the intersection of feature selection methods in convolutional neural network models. Measurement 158:107703. https://doi.org/10.1016/j.measurement.2020.107703
Toğaçar M, Ergen B, Özyurt F (2020) Evrişimsel Sinir Ağı Modellerinde Özellik Seçim Yöntemlerini Kullanarak Çiçek Görüntülerinin Sınıflandırılması. Fırat Üniversitesi Mühendislik Bilimleri Dergisi 32(1): 47–56. https://doi.org/10.35234/fumbd.573630
Ismail A, Idris MYI, Ayub MN, Yee Por L (2018) Vision-based apple classification for smart manufacturing. Sensors 18(12):4353. https://doi.org/10.3390/s18124353
Turkoglu M, Hanbay D, Sengur A (2019) Multi-model LSTM-based convolutional neural networks for detection of apple diseases and pests. J Ambient Intell Human Comput. https://doi.org/10.1007/s12652-019-01591-w
Hu Z, Tang J, Zhang P, Jiang J (2020) Deep learning for the identification of bruised apples by fusing 3D deep features for apple grading systems. Mech Syst Signal Process 145:106922. https://doi.org/10.1016/j.ymssp.2020.106922
Shrivastava VK, Pradhan MK, Minz S, Thakur MP (2019) Rice plant disease classification using transfer learning of deep convolution neural network. In: International archives of the photogrammetry, remote sensing & spatial information sciences, pp 631–635. https://doi.org/10.5194/isprs-archives-XLII-3-W6-631-2019
Jiang F, Lu Y, Chen Y, Cai D, Li G (2020) Image recognition of four rice leaf diseases based on deep learning and support vector machine. Comput Electron Agric 179:105824. https://doi.org/10.1016/j.compag.2020.105824
Sethy PK, Barpanda NK, Rath AK, Behera SK (2020) Deep feature based rice leaf disease identification using support vector machine. Comput Electron Agric 175:105527. https://doi.org/10.1016/j.compag.2020.105527
Verma S, Chug A, Singh AP (2020) Application of convolutional neural networks for evaluation of disease severity in tomato plant. J Discrete Math Sci Cryptogr 23(1):273–282. https://doi.org/10.1080/09720529.2020.1721890
Venal MCA, Fajardo AC, Hernandez AA (2019) Plant stress classification for smart agriculture utilizing convolutional neural network-support vector machine. In: 2019 International Conference on ICT for Smart Society (ICISS), 1–5. https://doi.org/10.1109/ICISS48059.2019.8969799
Zhuang S, Wang P, Jiang B, Li M (2020) Learned features of leaf phenotype to monitor maize water status in the fields. Comput Electron Agric 172:105347. https://doi.org/10.1016/j.compag.2020.105347
Zhu H, Yang L, Fei J, Zhao L, Han Z (2021) Recognition of carrot appearance quality based on deep feature and support vector machine. Comput Electron Agric 186:106185. https://doi.org/10.1016/j.compag.2021.106185
Koklu M, Unlersen MF, Ozkan IA, Aslan MF, Sabanci K (2022) A CNN-SVM study based on selected deep features for grapevine leaves classification. Measurement 188:110425. https://doi.org/10.1016/j.measurement.2021.110425
Sardogan M, Tuncer A, Ozen Y (2018) Plant leaf disease detection and classification based on CNN with LVQ algorithm. In: 2018 3rd International conference on computer science and engineering (UBMK), Sarajevo, Bosnia and Herzegovina, pp 382–385. https://doi.org/10.1109/UBMK.2018.8566635
Türkoğlu M, Hanbay D (2019) Plant disease and pest detection using deep learning-based features. Turk J Electr Eng Comput Sci 27(3):1636–1651. https://doi.org/10.3906/elk-1809-181
Goodfellow I, Lee H, Le Q, Saxe A, Ng A (2009) Measuring invariances in deep networks. In: Advances in neural information processing systems, vol 22, pp 646–654
Borji A, Izadi S, Itti L,(2015) What can we learn about CNNs from a large scale controlled object dataset? arXiv preprint arXiv:151201320
Brosnan T, Sun D-W (2004) Improving quality inspection of food products by computer vision: a review. J Food Eng 61(1):3–16. https://doi.org/10.1016/S0260-8774(03)00183-3
Büyükarıkan B, Üncü İS, (2019) Bilgisayarli görü sistemleri için sistem tasarımı ve kontrolü. Selçuk Üniversitesi Mühendislik, Bilim Ve Teknoloji Dergisi, 7(1): 228–240. https://doi.org/10.15317/Scitech.2019.194.
Kludt C, Längle T, Beyerer J (2021) Light field illumination: problem-specific lighting adjustment. tm-Technisches Messen 88(6):330–341. https://doi.org/10.1515/teme-2021-0021
Tao Y (1996) Spherical transform of fruit images for on-line defect extraction of mass objects. Opt Eng 35(2):344–350. https://doi.org/10.1117/1.600902
Cubero S, Aleixos N, Moltó E, Gómez-Sanchis J, Blasco J (2011) Advances in machine vision applications for automatic inspection and quality evaluation of fruits and vegetables. Food Bioprocess Technol 4(4):487–504. https://doi.org/10.1007/s11947-010-0411-8
Çalhan Ö (2014) Elmalarda Görülen Bazı Fizyolojik Bozukluklar, https://arastirma.tarimorman.gov.tr/marem/Belgeler/Yeti%C5%9Ftiricilik%20Bilgileri/Fizyolojik%20Bozukluklar.pdf. 6 March 2022
Nara K, Kato Y, Motomura Y (2001) Involvement of terminal-arabinose and-galactose pectic compounds in mealiness of apple fruit during storage. Postharvest Biol Technol 22(2):141–150. https://doi.org/10.1016/S0925-5214(00)00193-9
Unay D, Gosselin B (2007) Stem and calyx recognition on ‘Jonagold’apples by pattern recognition. J Food Eng 78(2):597–605. https://doi.org/10.1016/j.jfoodeng.2005.10.038
Hassan SM, Jasinski M, Leonowicz Z, Jasinska E, Maji AK (2021) Plant disease identification using shallow convolutional neural network. Agronomy 11(12):2388. https://doi.org/10.3390/agronomy11122388
Aqel D, Al-Zubi S, Mughaid A, Jararweh Y (2021) Extreme learning machine for plant diseases classification: a sustainable approach for smart agriculture. Cluster Computing, 1–14. https://doi.org/10.1007/s10586-021-03397-y
Al Bashish D, Braik M, Bani-Ahmad S (2011) Detection and classification of leaf diseases using K-means-based segmentation and neural-networks-based classification. Inf Technol J 10(2):267–275. https://doi.org/10.3923/itj.2011.267.275
Kulkarni AH, Patil A (2012) Applying image processing technique to detect plant diseases. Int J Mod Eng Res 2(5):3661–3664
Huang K, Aviyente S (2006) Sparse representation for signal classification. Adv Neural Inf Process Syst, 19
Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A (2015) Going deeper with convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition, Boston, Massachusetts, pp 1–9. https://doi.org/10.1109/CVPR.2015.7298594
Huang G, Liu Z, van der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, Honolulu, Hawaii, pp 4700–4708. https://doi.org/10.1109/CVPR.2017.243
Cetinic E, Lipic T, Grgic S (2018) Fine-tuning convolutional neural networks for fine art classification. Expert Syst Appl 114:107–118. https://doi.org/10.1016/j.eswa.2018.07.026
Bansal P, Kumar R, Kumar S (2021) Disease detection in apple leaves using deep convolutional neural network. Agriculture 11(7):617. https://doi.org/10.3390/agriculture11070617
Choi H-H, Yun B-J (2019) Illumination estimation for color constancy using convolutional neural network (CNN). Int J Signal Process 4:6–8
Ghazi MM, Yanikoglu B, Aptoula E (2017) Plant identification using deep neural networks via optimization of transfer learning parameters. Neurocomputing 235:228–235. https://doi.org/10.1016/j.neucom.2017.01.018
Al Mufti M, Al Hadhrami E, Taha B, Werghi N (2018) Automatic target recognition in SAR images: comparison between pre-trained CNNs in a transfer learning based approach. In: 2018 International conference on artificial intelligence and big data (ICAIBD), Chengdu, China, pp 160–164. https://doi.org/10.1109/ICAIBD.2018.8396186
Kim H, Lee H, Kim J-S, Ahn S-H (2020) Image-based failure detection for material extrusion process using a convolutional neural network. Int J Adv Manuf Technol 111(5):1291–1302. https://doi.org/10.1007/s00170-020-06201-0
Dyrmann M, Karstoft H, Midtiby HS (2016) Plant species classification using deep convolutional neural network. Biosyst Eng 151:72–80. https://doi.org/10.1016/j.biosystemseng.2016.08.024
Mohanty SP, Hughes DP, Salathé M (2016) Using deep learning for image-based plant disease detection. Front Plant Sci 7:1419. https://doi.org/10.3389/fpls.2016.01419
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:14091556
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, Las Vegas, 770–778. https://doi.org/10.1109/CVPR.2016.90
Zhang P, Yang L, Li D (2020) EfficientNet-B4-Ranger: a novel method for greenhouse cucumber disease recognition under natural complex environment. Comput Electron Agric 176:105652. https://doi.org/10.1016/j.compag.2020.105652
Cortes C, Vapnik V (1995) Support-vector networks. Machine Learning 20(3):273–297. https://doi.org/10.1007/BF00994018
Özkan Y (2016) Veri madenciligi yöntemleri. İstanbul, Turkey, Papatya Yayıncılık
Breiman L (2001) Random forests. Mach Learn 45(1):5–32. https://doi.org/10.1023/A:1010933404324
Qi Y (2012) Random forest for bioinformatics. In: Zhang C, Ma Y (eds) Ensemble machine learning. vol 1. Springer, Boston, pp 307–323. https://doi.org/10.1007/978-1-4419-9326-7_11
Koyuncugil AS, Özgülbaş N (2009) Veri madenciliği: Tıp ve sağlık hizmetlerinde kullanımı ve uygulamaları. Bilişim Teknolojileri Dergisi 2(2):21–32
Demir H, Erdoğmuş P, Kekeçoğlu M, (2018) Destek Vektör Makineleri, YSA, K-Means ve KNN Kullanarak Arı Türlerinin Sınıflandırılması. Düzce Üniversitesi Bilim ve Teknoloji Dergisi, 6 (1): 47–67. https://doi.org/10.29130/dubited.328596
Nijhawan R, Raman B, Das J (2018) Proposed hybrid-classifier ensemble algorithm to map snow cover area. J Appl Remote Sens 12(1):016003. https://doi.org/10.1117/1.JRS.12.016003
Chen T, Guestrin C, (2016) Xgboost: A scalable tree boosting system. In: KDD '16: Proceedings of the 22nd acm sigkdd international conference on knowledge discovery and data mining, San Francisco California USA, 785–794. https://doi.org/10.1145/2939672.2939785
Zhang H, Qiu D, Wu R, Deng Y, Ji D, Li T (2019) Novel framework for image attribute annotation with gene selection XGBoost algorithm and relative attribute model. Appl Soft Comput 80:57–79. https://doi.org/10.1016/j.asoc.2019.03.017
Anagnostis A, Asiminari G, Papageorgiou E, Bochtis D (2020) A convolutional neural networks based method for anthracnose infected walnut tree leaves identification. Appl Sci 10(2):469. https://doi.org/10.3390/app10020469
Funding
This work was supported by the Scientific Research Project at Konya Technical University, Konya, Turkey (No. 201113006).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Buyukarikan, B., Ulker, E. Classification of physiological disorders in apples fruit using a hybrid model based on convolutional neural network and machine learning methods. Neural Comput & Applic 34, 16973–16988 (2022). https://doi.org/10.1007/s00521-022-07350-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-022-07350-x