Abstract
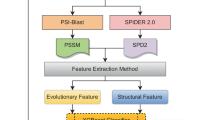
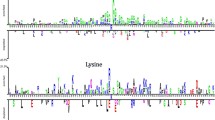
Post Translational Modification (PTM) plays an essential role in the biological and molecular mechanisms. They are also considered as a vital element in cell signaling and networking pathways. Among different PTMs, Methylation is regarded as one of the most important types. Methylation plays a crucial role in maintaining the dynamic balance, stability, and remodeling of chromatins. Methylation also leads to different abnormalities in cells and is responsible for many serious diseases. Methylation can be detected by experimental approaches such as methylation-specific antibodies, mass spectrometry, or characterizing methylation sites using the radioactive labeling method. However, these approaches are time-consuming and costly. Therefore, there is a demand for fast and accurate computational techniques to solve these issues. This study proposes a novel machine learning approach called MethEvo to predict methylation sites in proteins. To build this model, we use an evolutionary-based bi-gram profile approach to extract features. We also use SVM as our classification technique to build MethEvo. Our results demonstrate that MethEvo achieves 98.7%, 98.8%, 98.4%, and 0.974 in terms of accuracy, specificity, sensitivity, and Matthews Correlation Coefficient (MCC). MethEvo and its source code are publicly available at: https://github.com/islamsadia88/MethEvo.





Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Data availability
MethEvo and its source code are publicly available at: https://github.com/islamsadia88/MethEvo.
References
Cornett EM, Ferry L, Defossez PA, Rothbart SB (2019) Lysine methylation regulators moonlighting outside the epigenome. Mol Cell 75(6):1092–1101
Qiu WR, Xiao X, Lin WZ, Chou KC (2014) IMethyl-PseAAC: Identification of protein methylation sites via a pseudo amino acid composition approach. Biomed Res Int. https://doi.org/10.1155/2014/947416
Qiu H, Guo Y, Yu L, Pu X, Li M (2018) Predicting protein lysine methylation sites by incorporating single-residue structural features into Chou’s pseudo components. Chemom Intell Lab Syst 179:31–38
Cao XJ, Arnaudo AM, Garcia BA (2013) Large-scale global identification of protein lysine methylation in vivo. Epigenetics 8(5):477–485
Shien DM, Lee TY, Chang WC, Hsu JBK, Horng JT, Hsu PC, Wang TY, Huang HD (2009) Incorporating structural characteristics for identification of protein methylation sites. J Comput Chem 30(9):1532–1543
Liu H, Galka M, Mori E, Liu X, Lin YF, Wei R, Pittock P, Voss C, Dhami G, Li X, Miyaji M (2013) A method for systematic mapping of protein lysine methylation identifies functions for HP1β in DNA damage response. Mol Cell 50(5):723–735
Biggar KK, Charih F, Liu H, Ruiz-Blanco YB, Stalker L, Chopra A, Connolly J, Adhikary H, Frensemier K, Galka M, Fang Q (2020) Proteome-wide prediction of lysine methylation reveals novel histone marks and outlines the methyllysine proteome. Biorxiv. https://doi.org/10.1101/274688
Chen H, Xue Y, Huang N, Yao X, Sun Z (2006) MeMo: a web tool for prediction of protein methylation modifications. Nucleic Acid Res 34(suppl 2):W249–W253
Shao J, Xu D, Tsai SN, Wang Y, Ngai SM (2009) Computational identification of protein methylation sites through bi-profile Bayes feature extraction. PLoS One 4(3):e4920
Shi SP, Qiu JD, Sun XY, Suo SB, Huang SY, Liang RP (2012) PMeS: prediction of methylation sites based on enhanced feature encoding scheme. PLoS One 7(6):e38772
Wei L, Xing P, Shi G, Ji ZL, Zou Q (2017) Fast prediction of protein methylation sites using a sequence-based feature selection technique. IEEE/ACM Trans. Comput. Biol Bioinform 16:1–12
Zheng W, Wuyun Q, Cheng M, Hu G, Zhang Y (2020) Two-level protein methylation prediction using structure model-based features. Sci Rep 10(1):1–15
Ahmad MW, Arafat ME, Taherzadeh G, Sharma A, Dipta SR, Dehzangi A, Shatabda S (2020) Mal-light: enhancing lysine malonylation sites prediction problem using evolutionary-based features. IEEE Access 8:77888–77902
Shatabda S, Saha S, Sharma A, Dehzangi A (2017) iPHLoc-ES: identification of bacteriophage protein locations using evolutionary and structural features. J Theor Biol 435:229–237
Uddin MR, Sharma A, Farid DM, Rahman MM, Dehzangi A, Shatabda S (2018) EvoStruct-Sub: an accurate gram-positive protein subcellular localization predictor using evolutionary and structural features. J Theor Biol 443:138–146
Dehzangi A, López Y, Lal SP, Taherzadeh G, Michaelson J, Sattar A, Tsunoda T, Sharma A (2017) PSSM-Suc: accurately predicting succinylation using position specific scoring matrix into bigram for feature extraction. J Theor Biol 425:97–102
Liu Z, Wang Y, Gao T, Pan Z, Cheng H, Yang Q, Cheng Z, Guo A, Ren J, Xue Y (2014) CPLM: a database of protein lysine modifications. Nucleic Acid Res 42(D1):D531–D536
Reddy HM, Sharma A, Dehzangi A, Shigemizu D, Chandra AA, Tsunoda T (2019) GlyStruct: glycation prediction using structural properties of amino acid residues. BMC Bioinform. https://doi.org/10.1186/s12859-018-2547-x
Abid H, Jenny NJ, and Shovan SM (2020) Improved identification performance of lysine glycation PTM using PSI-BLAST. 2020 IEEE region 10 symposium TENSYMP 2020, pp 18–21
Xu Y, Ding YX, Ding J, Lei YH, Wu LY, Deng NY (2015) ISuc-PseAAC: Predicting lysine succinylation in proteins by incorporating peptide position-specific propensity. Sci Rep 5(June):3–8
Jia J, Liu Z, Xiao X, Liu B, Chou KC (2016) ISuc-PseOpt: identifying lysine succinylation sites in proteins by incorporating sequence-coupling effects into pseudo components and optimizing imbalanced training dataset. Anal Biochem 497:48–56
Fu L, Niu B, Zhu Z, Wu S, Li W (2012) CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28(23):3150–3152
Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP (2002) SMOTE: synthetic minority over-sampling technique. J Artif Intell Res 16:321–357
He H, Bai Y, Garcia EA, Li S (2008) ADASYN: adaptive synthetic sampling approach for imbalanced learning. Proc Int Jt Conf Neural Netw 3:1322–1328
Dehzangi A, López Y, Lal SP, Taherzadeh G, Sattar A, Tsunoda T, Sharma A (2018) Improving succinylation prediction accuracy by incorporating the secondary structure via helix, strand and coil, and evolutionary information from profile bigrams. PLoS One 13(2):e0191900
Islam MM, Saha S, Rahman MM, Shatabda S, Farid DM, Dehzangi A (2018) iProtGly-SS: identifying protein glycation sites using sequence and structure based features. Protein Struct Funct Bioinform 86(7):777–789
Heffernan R, Paliwal K, Lyons J, Dehzangi A, Sharma A, Wang J, Sattar A, Yang Y, Zhou Y (2015) Improving prediction of secondary structure, local backbone angles and solvent accessible surface area of proteins by iterative deep learning. Sci Rep 5(1):1–11
Sharma A, Lyons J, Dehzangi A, Paliwal KK (2013) A feature extraction technique using bi-gram probabilities of position specific scoring matrix for protein fold recognition. J Theor Biol 320:41–46
Noble WS (2006) What is a support vector machine? Nat Biotechnol 24(12):1565–1567
Patle A and Chouhan DS (2013) SVM kernel functions for classification. In 2013 international conference on advances in technology and engineering (ICATE), pp 1–9
Cai CZ, Han LY, Ji ZL, Chen X, Chen YZ (2003) SVM-Prot: web-based support vector machine software for functional classification of a protein from its primary sequence. Nucleic Acid Res 31(13):3692–3697
Lewis DP, Jebara T, Noble WS (2006) Support vector machine learning from heterogeneous data: an empirical analysis using protein sequence and structure. Bioinformatics 22(22):2753–2760
Guo Y, Yu L, Wen Z, Li M (2008) Using support vector machine combined with auto covariance to predict protein–protein interactions from protein sequences. Nucleic Acid Res 36(9):3025–3030
Kleinbaum DG (1994) Introduction to Logistic Regression. Springer, New York
Myles AJ, Feudale RN, Liu Y, Woody NA, Brown SD (2004) An introduction to decision tree modeling. J Chemom A J Chemome Soc 18(6):275–285
Natekin A, Knoll A (2013) Gradient boosting machines, a tutorial. Front Neurorobot 7:21
Jahromi AH and Taheri M (2017) A non-parametric mixture of gaussian naive bayes classifiers based on local independent features. In: 2017 artificial intelligence and signal processing conference (AISP) IEEE pp 209–212
Schapire RE (2013) Explaining adaboost. Empirical inference. Springer, Berlin, Heidelberg, pp 37–52
Biau G, Scornet E (2016) A random forest guided tour. Test 25(2):197–227
Davis J, Goadrich M (2006) The relationship between PR and ROC curves. ACM Int Conf Proc Ser 148:233–240
Chou K-C, Shen H-B (2009) REVIEW: recent advances in developing web-servers for predicting protein attributes. Nat Sci 01(02):63–92
Alinejad-Rokny H, Ghavami Modegh R, Rabiee HR, Ramezani Sarbandi E, Rezaie N, Tam KT, Forrest AR (2022) MaxHiC: a robust background correction model to identify biologically relevant chromatin interactions in Hi-C and capture Hi-C experiments. PLoS Comput Biol 18(6):e1010241
Dashti H, Dehzangi I, Bayati M, Breen J, Beheshti A, Lovell N (2022) Integrative analysis of mutated genes and mutational processes reveals novel mutational biomarkers in colorectal cancer. BMC Bioinform 23(1):1–24
Khakmardan S, Rezvani M, Pouyan AA, Fateh M (2020) MHiC, an integrated user-friendly tool for the identification and visualization of significant interactions in Hi-C data. BMC Genom 21(1):1–10
Javanmard R, JeddiSaravi K (2013) Proposed a new method for rules extraction using artificial neural network and artificial immune system in cancer diagnosis. J Bionanosci 7(6):665–672
Alinejad-Rokny H, Sadroddiny E, Scaria V (2018) Machine learning and data mining techniques for medical complex data analysis. Neurocomputing. https://doi.org/10.1016/j.neucom.2017.09.027
Niu H, Xu W, Akbarzadeh H, Parvin H, Beheshti A (2020) Deep feature learnt by conventional deep neural network. Comput Electr Eng 84:106656
Bayati M, Rabiee HR, Mehrbod M, Vafaee F, Ebrahimi D, Forrest AR (2020) CANCERSIGN: a user-friendly and robust tool for identification and classification of mutational signatures and patterns in cancer genomes. Sci Rep 10(1):1–11
Rajaei P, Jahanian KH, Beheshti A, Band SS, Dehzangi A (2021) VIRMOTIF: a user-friendly tool for viral sequence analysis. Genes 12(2):186
Sharifrazi D, Alizadehsani R, Joloudari JH, Shamshirband S, Hussain S, Sani ZA (2022) CNN-KCL: automatic myocarditis diagnosis using convolutional neural network combined with k-means clustering. Math Biosci Eng 19(3):2381–2402
Funding
This research received no external funding Rutgers, The State University of New Jersey, p321243, Abdollah Dehzangi.
Author information
Authors and Affiliations
Contributions
Conceptualization, I.D., H.A.R and S.S.; methodology, S.I, S.B.S.M, S.R.D, and MD.E.A software, S.I, R.R.D; validation, S.B.S.M, S.I. and S.S.; formal analysis, S.I.; investigation, I.D; resources, S.S., and I.D; data curation, I.D.; writing—original draft preparation, S.I., I.D, AND S.S; writing—review and editing, I.D, AND H.A.R; supervision, S.S., H.A.R, AND I.D.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Informed consent
Informed consent was obtained from all subjects involved in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Islam, S., Mugdha, S.B.S., Dipta, S.R. et al. MethEvo: an accurate evolutionary information-based methylation site predictor. Neural Comput & Applic 36, 201–212 (2024). https://doi.org/10.1007/s00521-022-07738-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-022-07738-9