Abstract
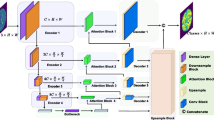
Delineating accurately and simultaneously all lesions is vital and challenging for computer-aided diagnosis for multiple neurofibromatosis (NF). However, existing CNN-based segmentation methods paid little attention to weak boundaries. Moreover, due to the intensity-inhomogeneous distribution of medical images, the ambiguous boundaries, and highly variable locations, sizes and shapes of the lesions, delineating multiple lesions simultaneously remains quite challenging. To address these challenges, we introduce a novel end-to-end segmentation framework of multiple NF, deep hierarchical geodesic active contour (DH-GAC). It leverages the elaborately designed deep hierarchical context fusion network (DH-CFN) to improve the generalization and robustness of DH-GAC, and the modified geodesic active contour (MGAC) to delineate precisely all lesions as much as possible. Specifically, it employs DH-CFN to predict specific parameter maps of each image for MGAC and feeds them into the energy function of MGAC to delineate NF lesions, which makes DH-GAC end-to-end trainable. Moreover, to improve the generalization of DH-GAC, we adopt two different settings to initialize the surface for DH-GAC. Experimental results demonstrate that DH-GAC not only improves the segmentation precision, but also overcomes the intrinsic drawback of classical geodesic active contour in boundary delineation.









Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Data availability
The data that support the findings of this study are available from Harvard Medical School but restrictions apply to the availability of these data, which were used under licence for the current study, and so are not publicly available. Data are however available from the last author upon reasonable request and with permission of Harvard Medical School.
References
Otsu N (1975) A threshold selection method from gray-level histogram. Automatica 11:285–296
Saleh S, Kalyankar NV, Khamitkar S (2010) Image segmentation by using threshold techniques. J Comput 2(5):2151–9617
Adams R, Bischof L (1994) Seeded region growing. IEEE Trans Pattern Anal Mach Intell 16:641–647. https://doi.org/10.1109/34.295913
Boykov YY, Jolly M-P (2001) Interactive graph cuts for optimal boundary region segmentation of objects in n-d images. In: Proceedings Eighth IEEE International Conference on Computer Vision. ICCV 2001, vol 1, pp 105–112. https://doi.org/10.1109/ICCV.2001.937505
Rother C, Kolmogorov V, Blake A (2004) Grabcut: interactive foreground extraction using iterated graph cuts. ACM Trans Graph 23:309–314. https://doi.org/10.1145/1186562.1015720
Felzenszwalb P, Huttenlocher D (2004) Efficient graph-based image segmentation. Int J Comput Vis 59:167–181. https://doi.org/10.1023/B:VISI.0000022288.19776.77
Kass M, Witkin A, Terzopoulos D (1988) Snakes: active contour models. Int J Comput Vis 1(4):321–331. https://doi.org/10.1016/B978-0-12-386454-3.00786-7
Cohen LD (1991) On active contour models and balloons. CVGIP: Image Underst 53(2):211–218
Xu C, Prince JL (1998) Snakes, shapes, and gradient vector flow. IEEE Trans Image Process 7(3):359–369. https://doi.org/10.1109/83.661186
Xu C, Prince J (2000) Generalized gradient vector flow external forces for active contours. Signal Process 71:131–139. https://doi.org/10.1016/S0165-1684(98)00140-6
Jifeng N, Chengke W, Shigang L, Shuqin Y (2007) NGVF: an improved external force field for active contour model. Pattern Recogn Lett 28(1):58–63. https://doi.org/10.1016/j.patrec.2006.06.014
Jia X, Wang Y (2009) An edge preserving gradient vector flow for active contours. In: Proceedings of the 2009 2nd International Congress on Image and Signal Processing, CISP’09. https://doi.org/10.1109/CISP.2009.5304695
Wang Y, Liu L, Zhang H, Cao Z, Lu S (2010) Image segmentation using active contours with normally biased gvf external force. IEEE Signal Process Lett 17(10):875–878. https://doi.org/10.1109/LSP.2010.2060482
Yao Y, Liu L, Liao L, Wei M, Guo J, Li Y (2012) Sigmoid gradient vector flow for medical image segmentation. Int Conf Signal Process Proc ICSP 2(2):881–884. https://doi.org/10.1109/ICoSP.2012.6491721
Osher S, Sethian J (1988) Fronts propagating with curvature dependent speed algorithms based on hamilton-jacobi. J Comput Phys 79:12–49. https://doi.org/10.1016/0021-9991(88)90002-2
Caselles V, Kimmel R, Sapiro G (1997) Geodesic active contours. Int J Comput Vis 22(1):61–79. https://doi.org/10.1023/A:1007979827043
Goldenberg R, Kimmel R, Rivlin E, Rudzsky M (2001) Fast geodesic active contours. IEEE Trans Image Process 10(10):1467–1475. https://doi.org/10.1109/83.951533
Paragios N, Mellina-Gottardo O, Ramesh V (2004) Gradient vector flow fast geometric active contours. IEEE Trans Pattern Anal Mach Intell 26(3):402–407. https://doi.org/10.1109/TPAMI.2004.1262337
Chan TF, Vese LA (2001) Active contours without edges. IEEE Trans Image Process 10(2):266–277. https://doi.org/10.1109/83.902291
Li C, Kao C, Gore JC, Ding Z (2007) Implicit active contours driven by local binary fitting energy. In: 2007 IEEE Conference on Computer Vision and Pattern Recognition, pp 1–7. https://doi.org/10.1109/CVPR.2007.383014
Feng C (2017) IEOPF: An active contour model for image segmentation with inhomogeneities estimated by orthogonal primary functions
Li C, Huang R, Ding Z, Gatenby JC, Metaxas DN, Gore JC (2011) A level set method for image segmentation in the presence of intensity inhomogeneities with application to mri. IEEE Trans Image Process 20(7):2007–2016. https://doi.org/10.1109/TIP.2011.2146190
Li X, Liu H, Xing Y (2019) A hybride active contour model driven by global and local image information. Neural Process Lett. https://doi.org/10.1007/s11063-019-10004-0
Tan G, Guo Z, Xiao Y (2019) PA-RetinaNet: path augmented retinaNet for dense object detection, pp. 138–149. https://doi.org/10.1007/978-3-030-30484-3_12
Chen C, Li K, Zou X, Zhongyao C, Wei W, Tian Q, Zeng Z (2021) Hierarchical semantic graph reasoning for train component detection. IEEE Trans Neural Netw Learn Syst. https://doi.org/10.1109/TNNLS.2021.3057792
Wu X, Tan G, Zhu N, Chen Z, Yang Y, Wen H, Li K (2021) Cachetrack-yolo: Real-time detection and tracking for thyroid nodules and surrounding tissues in ultrasound videos. IEEE J Biomed Health Inform 25(10):3812–3823. https://doi.org/10.1109/JBHI.2021.3084962
Shelhamer E, Long J, Darrell T (2017) Fully convolutional networks for semantic segmentation. IEEE Trans Pattern Anal Mach Intell 39(4):640–651. https://doi.org/10.1109/TPAMI.2016.2572683
Badrinarayanan V, Kendall A, Cipolla R (2017) Segnet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Transactions on Pattern Analysis and Machine Intelligence 39(12):2481–2495. https://doi.org/10.1109/TPAMI.2016.2644615
Ronneberger O, Fischer P, Brox T (2015) U-net: convolutional networks for biomedical image segmentation. In: Lecture notes in computer science (including subseries lecture notes in artificial intelligence and lecture notes in bioinformatics) 9351:234–241. https://doi.org/10.1007/978-3-319-24574-4_28
Chen L-C, Papandreou G, Kokkinos I, Murphy K, Yuille AL (2018) Deeplab: semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans Pattern Anal Mach Intell 40(4):834–848. https://doi.org/10.1109/TPAMI.2017.2699184
Zhou Z, Rahman Siddiquee MM, Tajbakhsh N, Liang J (2018) Unet++: A nested u-net architecture for medical image segmentation. In: Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support, pp 3–11. Springer, Cham. https://doi.org/10.1007/978-3-030-00889-5_1
He K, Gkioxari G, Dollár P, Girshick R (2017) Mask r-cnn. In: 2017 IEEE International Conference on Computer Vision (ICCV), pp. 2980–2988. https://doi.org/10.1109/ICCV.2017.322
Wu X, Tan G, Li K, Li S, Wen H, Zhu X, Cai W (2020) Deep parametric active contour model for neurofibromatosis segmentation. Future Gener Comput Syst 112:58–66. https://doi.org/10.1016/j.future.2020.05.001
Cai Z, Vasconcelos N (2021) Cascade r-cnn: high quality object detection and instance segmentation. IEEE Trans Pattern Anal Mach Intell 43(5):1483–1498. https://doi.org/10.1109/TPAMI.2019.2956516
Peng S, Jiang W, Pi H, Li X, Bao H, Zhou X (2020) Deep snake for real-time instance segmentation. In: 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pp. 8530–8539. https://doi.org/10.1109/CVPR42600.2020.00856
Ranjbarzadeh R, Bagherian Kasgari A, Jafarzadeh Ghoushchi S, Anari S, Naseri M, Bendechache M (2021) Brain tumor segmentation based on deep learning and an attention mechanism using mri multi-modalities brain images. Sci Rep 11:567–577. https://doi.org/10.1038/s41598-021-90428-8
Jin Q, Cui H, Sun C, Meng Z, Su R (2021) Cascade knowledge diffusion network for skin lesion diagnosis and segmentation. Appl Soft Comput 99:106881. https://doi.org/10.1016/j.asoc.2020.106881
Chen C, Li K, Wei W, Zhou J, Zeng Z (2021) Hierarchical graph neural networks for few-shot learning. IEEE Trans Circ Syst Video Technol. https://doi.org/10.1109/TCSVT.2021.3058098
Pu B, Zhu N, Li K, Li S (2021) Fetal cardiac cycle detection in multi-resource echocardiograms using hybrid classification framework. Future Gener Comput Syst 115:825–836. https://doi.org/10.1016/j.future.2020.09.014
Pu B, Li K, Li S, Zhu N (2021) Automatic fetal ultrasound standard plane recognition based on deep learning and iiot. IEEE Trans Ind Inf 17(11):7771–7780. https://doi.org/10.1109/TII.2021.3069470
Chen J, Li K, Li K, Yu P, Zeng Z (2021) Dynamic planning of bicycle stations in dockless public bicycle-sharing system using gated graph neural network. ACM Trans Intell Syst Technol 12:1–22. https://doi.org/10.1145/3446342
Chen J, Li K, Tang Z, Bilal K, Yu S, Weng C, Li K (2017) A parallel random forest algorithm for big data in a spark cloud computing environment. IEEE Trans Parallel Distrib Syst 28(4):919–933. https://doi.org/10.1109/TPDS.2016.2603511
Rupprecht C, Huaroc E, Baust M, Navab N (2016) Deep active contours. https://arxiv.org/abs/1607.05074
Chen H, Qi X, Yu L, Heng P-A (2016) Dcan: Deep contour-aware networks for accurate gland segmentation. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 2487–2496. https://doi.org/10.1109/CVPR.2016.273
Bai M, Urtasun R (2017) Deep watershed transform for instance segmentation. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 2858–2866. https://doi.org/10.1109/CVPR.2017.305
Kristiadi A (2017) Pranowo: deep convolutional level set method for image segmentation. J ICT Res Appl 11(3):284–298. https://doi.org/10.5614/itbj.ict.res.appl.2017.11.3.5
Hu P, Shuai B, Liu J, Wang G (2017) Deep level sets for salient object detection. In: Proceedings - 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017, pp 540–549. https://doi.org/10.1109/CVPR.2017.65
Marcos D, Tuia D, Kellenberger B, Zhang L, Bai M, Liao R, Urtasun R (2018) Learning deep structured active contours end-to-end. In: Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, pp 8877–8885. https://doi.org/10.1109/CVPR.2018.00925
Cheng D, Liao R, Fidler S, Urtasun R (2019) Darnet: Deep active ray network for building segmentation. In: 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pp 7423–7431. https://doi.org/10.1109/CVPR.2019.00761
Cerrone L, Zeilmann A, Hamprecht FA (2019) End-to-end learned random walker for seeded image segmentation. In: 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pp 12551–12560. https://doi.org/10.1109/CVPR.2019.01284
Hatamizadeh A, Hoogi A, Sengupta D, Lu W, Wilcox B, Rubin D, Terzopoulos D (2019) Deep active lesion segmentation. In: Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), pp 98–105. https://doi.org/10.1101/794529
Hatamizadeh A, Sengupta D, Terzopoulos D (2020) End-to-end trainable deep active contour models for automated image segmentation: delineating buildings in aerial imagery. In: Computer Vision – ECCV 2020, pp 730–746. https://doi.org/10.1007/978-3-030-58610-2_43
Chen X, Williams BM, Vallabhaneni SR, Czanner G, Williams R, Zheng Y (2019) Learning active contour models for medical image segmentation. In: 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pp 11624–11632. https://doi.org/10.1109/CVPR.2019.01190
Kim Y, Kim S, Kim T, Kim C (2019) CNN-based semantic segmentation using level set loss. In: Proceedings - 2019 IEEE Winter Conference on Applications of Computer Vision, WACV 2019, pp 1752–1760. https://doi.org/10.1109/WACV.2019.00191
Kim B, Ye JC (2020) Mumford-shah loss functional for image segmentation with deep learning. IEEE Trans Image Process 29:1856–1866. https://doi.org/10.1109/TIP.2019.2941265
Zhao X, Yin Y, Yang B, Zhu B, Tian X (2007) Level set and geodesic active contours based measurement of material removal between serial sections. Comput Mater Sci 39:857–861. https://doi.org/10.1016/j.commatsci.2006.10.018
Caselles V, Catte F, Coll B, Dibos F (1993) A geometric model for active contours in image processing. Numer Math 66:1–31. https://doi.org/10.1007/BF01385685
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 62072168) and Natural Science Foundation of Hunan Province (Grant No. 2021JJ30148).
Author information
Authors and Affiliations
Contributions
Conceptualization was contributed by XW, GT, and WC; methodology was contributed by GT and WC; formal analysis and investigation were contributed by XW; technical guidance was contributed by GT and WC; writing-original draft preparation was contributed by XW; writing-review and editing was contributed by BP and MD; funding acquisition was contributed by GT and WC; visualization was contributed by XW and BP; and resources were contributed by MD and WC.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this article.
Ethical approval
The article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wu, X., Tan, G., Pu, B. et al. DH-GAC: deep hierarchical context fusion network with modified geodesic active contour for multiple neurofibromatosis segmentation. Neural Comput & Applic 37, 7511–7526 (2025). https://doi.org/10.1007/s00521-022-07945-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-022-07945-4