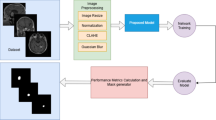
Abstract
Recently, U-Net architecture with its strong adaptability has become prevalent in the field of MRI brain tumor segmentation. Meanwhile, researchers have demonstrated that introducing attention mechanisms, especially self-attention, into U-Net can effectively improve the performance of segmentation models. However, the self-attention has disadvantages of heavy computational burden, quadratic complexity as well as ignoring the potential correlations between different samples. Besides, current attention segmentation models seldom focus on adaptively computing the receptive field of tumor images that may capture discriminant information effectively. To address these issues, we propose a novel 3D U-Net related brain tumor segmentation model dubbed as self-calibrated attention U-Net (SCAU-Net) in this work, which simultaneously introduces two lightweight modules, i.e., external attention module and self-calibrated convolution module, into a single U-Net. More specifically, SCAU-Net embeds the external attention into the skip connection to better utilize encoding features for semantic up-sampling, and it leverages several 3D self-calibrated convolution modules to replace the original convolution layers, which adaptively computes the receptive field of tumor images for effective segmentation. SCAU-Net achieves segmentation results on the BraTS 2020 validation dataset with the dice similarity coefficient of 0.905, 0.821 and 0.781 and the 95% Hausdorff distance (HD95) of 4.0, 9.7 and 29.3 on the whole tumor, tumor core and enhancing tumor, respectively. Similarly, competitive results are obtained on BraTS 2018 and BraTS 2019 validation datasets. Experimental results demonstrate that SCAU-Net outperforms its baseline and achieves outstanding performance compared to various representative brain tumor models.





Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Availability of data and materials
The datasets during the current study are available in the: https://www.cbica.upenn.edu/MICCAI_BraTS2020_TrainingDatahttps://www.cbica.upenn.edu/MICCAI_BraTS2020_ValidationDatahttps://www.cbica.upenn.edu/sbia/Spyridon.Bakas/MICCAI_BraTS/2019/MICCAI_BraTS_2019_Data_Training.ziphttps://www.cbica.upenn.edu/sbia/Spyridon.Bakas/MICCAI_BraTS/2019/MICCAI_BraTS_2019_Data_Validation.zip.
References
Miller KD, Ostrom QT, Kruchko C, Patil N, Tihan T, Cioffi G, Fuchs HE, Waite KA, Jemal A, Siegel RL et al. (2021) Brain and other central nervous system tumor statistics, 2021. CA Cancer Journal Clin 71(5):381–406
Liu J, Li M, Wang J, Wu F, Liu T, Pan Y (2014) A survey of mri-based brain tumor segmentation methods. Tsinghua Sci Technol 19(6):578–595
Chahal PK, Pandey S (2021) A hybrid weighted fuzzy approach for brain tumor segmentation using mr images. Neural Comput Appl, pp 1–15
Amin J, Anjum MA, Gul N, Sharif M (2022) A secure two-qubit quantum model for segmentation and classification of brain tumor using mri images based on blockchain. Neural Comput Appl, pp 1–14
Liu P, Dou Q, Wang Q, Heng PA (2020) An encoder-decoder neural network with 3d squeeze-and-excitation and deep supervision for brain tumor segmentation. IEEE Access 8:34029–34037
Hu J, Shen L, Sun G (2018) Squeeze-and-excitation networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 7132–7141
Zhu Z, Xu M, Bai S, Huang T, Bai X (2019) Asymmetric non-local neural networks for semantic segmentation. In: Proceedings of the IEEE/CVF international conference on computer vision, pp 593–602
Jia H, Cai W, Huang H, Xia Y (2021) H2nf-net for brain tumor segmentation using multimodal mr imaging: 2nd place solution to brats challenge 2020 segmentation task. In: Brainlesion: glioma, multiple sclerosis, stroke and traumatic brain injuries-6th international workshop, Springer, pp 58–68
Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser Ł, Polosukhin I (2017) Attention is all you need. Adv Neural Inf Process Syst, 30
Chen J, Lu Y, Yu Q, Luo X, Adeli E, Wang Y, Lu L, Yuille AL, Zhou Y (2021) Transunet: Transformers make strong encoders for medical image segmentation. Preprint at arXiv:2102.04306
Wang W, Chen C, Ding M, Yu H, Zha S, Li J (2021) Transbts: Multimodal brain tumor segmentation using transformer. In: International conference on medical image computing and computer-assisted intervention, pp 109–119. Springer
Valanarasu JMJ, Oza P, Hacihaliloglu I, Patel VM (2021) Medical transformer: gated axial-attention for medical image segmentation. In: Medical Image computing and computer assisted intervention–MICCAI 2021: 24th international conference, Strasbourg, France, 27 Sep–1 Oct, 2021, Proceedings, Part I 24, pp 36–46. Springer
Liu H, Zhang J, Yang K, Hu X, Stiefelhagen R (2022) Cmx: Cross-modal fusion for rgb-x semantic segmentation with transformers. arXiv preprint arXiv:2203.04838
Guo MH, Liu ZN, Mu TJ, Hu SM (2022) Beyond self-attention: external attention using two linear layers for visual tasks. IEEE Trans Pattern Anal Mach Intell 45(5):5436–5447
Liu JJ, Hou Q, Cheng MM, Wang C, Feng J (2020) Improving convolutional networks with self-calibrated convolutions. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 10096–10105
Wang X, Girshick R, Gupta A, He K (2018) Non-local neural networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 7794–7803
Fu J, Liu J, Tian H, Li Y, Bao Y, Fang Z, Lu H (2019) Dual attention network for scene segmentation. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 3146–3154
Zhang H, Goodfellow I, Metaxas D, Odena A (2019) Self-attention generative adversarial networks. In: International conference on machine learning, pp 7354–7363. PMLR
Yuan Y, Huang L, Guo J, Zhang C, Chen X, Wang J (2018) Ocnet: Object context network for scene parsing. Preprint at arXiv:1809.00916
Bello I, Zoph B, Vaswani A, Shlens J, Le QV (2019) Attention augmented convolutional networks. In: Proceedings of the IEEE/CVF international conference on computer vision, pp 3286–3295
Menze BH, Jakab A, Bauer S, Kalpathy Cramer J, Farahani K, Kirby J, Burren Y, Porz N, Slotboom J, Wiest R et al (2014) The multimodal brain tumor image segmentation benchmark (brats). IEEE Trans Med Imag 34(10):1993–2024
Bakas S, Akbari H, Sotiras A, Bilello M, Rozycki M, Kirby JS, Freymann JB, Farahani K, Davatzikos C (2017) Advancing the cancer genome atlas glioma mri collections with expert segmentation labels and radiomic features. Sci Data 4(1):1–13
Bakas S, Reyes M, Jakab A, Bauer S, Rempfler M, Crimi A, Shinohara RT, Berger C, Ha SM, Rozycki M, et al (2018) Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the brats challenge. Preprint at arXiv:1811.02629
Rohlfing T, Zahr NM, Sullivan EV, Pfefferbaum A (2010) The sri24 multichannel atlas of normal adult human brain structure. Human Brain Map 31(5):798–819
Isensee F, Kickingereder P, Wick W, Bendszus M, Maier-Hein KH (2018) No new-net. In: International MICCAI brainlesion workshop, pp 234–244. Springer
Chen C, Liu X, Ding M, Zheng J, Li J (2019) 3d dilated multi-fiber network for real-time brain tumor segmentation in mri. In: International conference on medical image computing and computer-assisted intervention, pp 184–192. Springer
Brügger R, Baumgartner CF, Konukoglu E (2019) A partially reversible u-net for memory-efficient volumetric image segmentation. In: International conference on medical image computing and computer-assisted intervention, pp 429–437. Springer
Zhou C, Ding C, Wang X, Lu Z, Tao D (2020) One-pass multi-task networks with cross-task guided attention for brain tumor segmentation. IEEE Trans Image Process 29:4516–4529
Myronenko A (2018) 3d mri brain tumor segmentation using autoencoder regularization. In: International MICCAI brainlesion workshop, pp 311–320. Springer
Elhamzi W, Ayadi W, Atri M (2022) A novel automatic approach for glioma segmentation. Neural Comput Appl 34(22):20191–20201
Kao PY, Ngo T, Zhang A, Chen JW, Manjunath B (2018) Brain tumor segmentation and tractographic feature extraction from structural mr images for overall survival prediction. In: International MICCAI brainlesion workshop, pp 128–141. Springer
Zhao YX, Zhang YM, Liu CL (2020) Bag of tricks for 3d mri brain tumor segmentation. In: International MICCAI brainlesion workshop, pp 210–220. Springer
Li X, Luo G, Wang K (2020) Multi-step cascaded networks for brain tumor segmentation. In: International MICCAI brainlesion workshop, pp 163–173. Springer
Chen M, Wu Y, Wu J (2020) Aggregating multi-scale prediction based on 3d u-net in brain tumor segmentation. In: International MICCAI brainlesion workshop, pp 142–152. Springer
Cheng X, Jiang Z, Sun Q, Zhang J (2020) Memory-efficient cascade 3d u-net for brain tumor segmentation. In: International MICCAI brainlesion workshop, pp 242–253. Springer
Guo D, Wang L, Song T, Wang G (2020) Cascaded global context convolutional neural network for brain tumor segmentation. In: International MICCAI brainlesion workshop, pp 315–326. Springer
Zhou T, Canu S, Vera P, Ruan S (2021) Latent correlation representation learning for brain tumor segmentation with missing mri modalities. IEEE Trans Image Process 30:4263–4274
Ahmad P, Jin H, Qamar S, Zheng R, Saeed A (2021) Rd2a: densely connected residual networks using aspp for brain tumor segmentation. Multimedia Tools Appl 80(18):27069–27094
Tang J, Li T, Shu H, Zhu H (2021) Variational-autoencoder regularized 3d multiresunet for the brats 2020 brain tumor segmentation. In: International MICCAI brainlesion workshop, pp 431–440. Springer
Cheng K, Hu C, Yin P, Su Q, Zhou G, Wu X, Wang X, Yang W (2021) Glioma sub-region segmentation on multi-parameter mri with label dropout. In: International MICCAI brainlesion workshop, pp 420–430. Springer
Sundaresan V, Griffanti L, Jenkinson M (2021) Brain tumour segmentation using a triplanar ensemble of u-nets on mr images. In: International MICCAI brainlesion workshop, pp 340–353. Springer
Guan X, Yang G, Ye J, Yang W, Xu X, Jiang W, Lai X (2022) 3d agse-vnet: an automatic brain tumor mri data segmentation framework. BMC Med Imag 22(1):1–18
Fang Y, Huang H, Yang W, Xu X, Jiang W, Lai X (2022) Nonlocal convolutional block attention module vnet for gliomas automatic segmentation. Int J Imag Syst Technol 32(2):528–543
Huang H, Yang G, Zhang W, Xu X, Yang W, Jiang W, Lai X (2021) A deep multi-task learning framework for brain tumor segmentation. Front Oncol 11:690244
Acknowledgements
This work was partially supported by the National Natural Science Foundation of China under Grant 61972062, the NSFC-Liaoning Province United Foundation under Grant U1908214 and the Young and Middle-aged Talents Program of the National Civil Affairs Commission.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, D., Sheng, N., Han, Y. et al. SCAU-net: 3D self-calibrated attention U-Net for brain tumor segmentation. Neural Comput & Applic 35, 23973–23985 (2023). https://doi.org/10.1007/s00521-023-08872-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-023-08872-8