Abstract
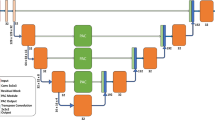
Segmentation of the liver from abdominal CT images is an essential step for computer-aided diagnosis and surgery planning. The U-Net architecture is one of the most well-known CNN architectures which achieved remarkable successes in both medical and biological image segmentation domain. However, it does not perform well when the target area is small or partitioned. In this paper, we propose a novel architecture, called dense feature selection U-Net (DFS U-Net), which addresses this challenging problem. Specifically, The Hounsfield unit values were windowed in a range to exclude irrelevant organs, and then use the pre-processed data to train our proposed DFS U-Net model. To further improve the segmentation accuracy of the small region and disconnected regions of interests with limited training datasets, we improve the loss function by adding a parameter to the formula. With respect to the ground truth, the Dice score ratio can reach over 94.9% for the liver. Our experimental results demonstrate its potential in clinical usage with high effectiveness, robustness and efficiency.














Similar content being viewed by others
References
Yu, Q., Xie, L., Wang, Y., Zhou, Y., Fishman, E.K., Yuille, A.L.: Recurrent saliency transformation network: Incorporating multi-stage visual cues for small organ segmentation, In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 8280–8289 (2018)
Yang, L., Zhang, Y., Chen, J., Zhang, S., Chen, D.Z.: Suggestive annotation: a deep active learning framework for biomedical image segmentation. In: International conference on medical image computing and computer-assisted intervention, pp. 399–407 (2017)
Yang, X., Liu, C., Wang, Z., Yang, J., Le Min, H., Wang, L., Cheng, K.T.T.: Co-trained convolutional neural networks for automated detection of prostate cancer in multi-parametric MRI. In: Medical image analysis, pp. 212–227(2017)
Shelhamer, E., Long, J., Darrell, T.: Fully convolutional networks for semantic segmentation. In: IEEE transactions on pattern analysis and machine intelligence, pp. 640–651 (2017)
Chen, L.C., Papandreou, G., Kokkinos, I., Murphy, K., Yuille, A.L.: Semantic image segmentation with deep convolutional nets and fully connected crfs. arXiv:1412.7062 (2014)
Ronneberger, O., Fischer, P., Brox, T.: U-net: Convolutional networks for biomedical image segmentation. In: International conference on medical image computing and computer-assisted intervention, pp. 234–241 (2015)
Wang, Z., Liu, C., Cheng, D., Wang, L., Yang, X., Cheng, K.T.: Automated detection of clinically significant prostate cancer in mp-MRI images based on an end-to-end deep neural network. In: IEEE transactions on medical imaging, pp. 1127–1139 (2018)
Nie, D., Gao, Y., Wang, L., Shen, D.: ASDNet: attention based semi-supervised deep networks for medical image segmentation. In: International conference on medical image computing and computer-assisted intervention, pp. 370–378 (2018)
Zhang, Y., Yang, L., Chen, J., Fredericksen, M., Hughes, D.P., Chen, D.Z.: Deep adversarial networks for biomedical image segmentation utilizing unannotated images. In: International conference on medical image computing and computer-assisted intervention, pp. 408–416 (2017)
Yang, D., Xu, D., Zhou, S.K., Georgescu, B., Chen, M., Grbic, S., Comaniciu, D.: Automatic liver segmentation using an adversarial image-to-image network, In: International conference on medical image computing and computer-assisted intervention, pp. 507–515 (2017)
Zheng, S., Jayasumana, S., Romera-Paredes, B., Vineet, V., Su, Z., Du, D., Torr, P.H.: Conditional random fields as recurrent neural networks. In: Proceedings of the IEEE international conference on computer vision, pp. 1529–1537 (2015)
Yan, Z., Yang, X., Cheng, K.T.: A skeletal similarity metric for quality evaluation of retinal vessel segmentation. In: IEEE transactions on medical imaging, pp. 1045–1057 (2017)
Xie, S., Tu, Z.: Holistically-nested edge detection. In: Proceedings of the IEEE international conference on computer vision, pp. 1395–1403 (2015)
Roth, H.R., Lu, L., Lay, N., Harrison, A.P., Farag, A., Sohn, A., Summers, R.M.: Spatial aggregation of holistically-nested convolutional neural networks for automated pancreas localization and segmentation. In: Medical image analysis, pp. 94–107 (2018)
Brügger, R., Baumgartner, C.F., Konukoglu, E.: A partially reversible U-Net for memory-efficient volumetric image segmentation. In: International conference on medical image computing and computer-assisted intervention, pp. 429–437 (2019)
Huang, G., Liu, Z., Van Der Maaten, L., Weinberger, K.Q.: Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 4700–4708 (2017)
Zhang, H., Patel, V.M.: Densely connected pyramid dehazing network. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 3194–3203 (2018)
Hu, J., Shen, L., Sun, G.: Squeeze-and-excitation networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 7132–7141 (2018)
Dou, Q., Chen, H., Jin, Y., Yu, L., Qin, J., Heng, P.A.: 3D deeply supervised network for automatic liver segmentation from CT volumes. In: International conference on medical image computing and computer-assisted intervention, pp. 149–157 (2016)
Liao, M., Zhao, Y.Q., Wang, W., Zeng, Y.Z., Yang, Q., Shih, F.Y., Zou, B.J.: Efficient liver segmentation in CT images based on graph cuts and bottleneck detection. In: Physica Medica, pp. 1383–1396 (2016)
Yang, X., Yu, H.C., Choi, Y., Lee, W., Wang, B., Yang, J., You, H.: A hybrid semi-automatic method for liver segmentation based on level-set methods using multiple seed points. In: Computer methods and programs in biomedicine, pp. 69–79 (2014)
Dou, Q., Yu, L., Chen, H., Jin, Y., Yang, X., Qin, J., Heng, P.A.: 3D deeply supervised network for automated segmentation of volumetric medical images. Med. Image Anal. 2017, 40–54 (2017)
Huang, W., Yang, Y., Lin, Z., Huang, G.B., Zhou, J., Duan, Y., Xiong, W.: Random feature subspace ensemble based extreme learning machine for liver tumor detection and segmentation. In: 2014 36th annual international conference of the IEEE engineering in medicine and biology society, pp. 4675–4678 (2014)
Jin, X., Ye, H., Li, L., Xia, Q.: Image segmentation of liver CT based on fully convolutional network. In: 2017 10th international symposium on computational intelligence and design (ISCID), pp. 210–213 (2017)
Li, X., Chen, H., Qi, X., Dou, Q., Fu, C.W., Heng, P.A.: H-DenseUNet: hybrid densely connected UNet for liver and tumor segmentation from CT volumes. In: IEEE transactions on medical imaging, pp. 2663–2674 (2018)
Kaluva, K.C., Khened, M., Kori, A., Krishnamurthi, G.: 2d-densely connected convolution neural networks for automatic liver and tumor segmentation. arXiv:1802.02182 (2018)
Christ, P.F., Ettlinger, F., Grün, F., Elshaera, M.E.A., Lipkova, J., Schlecht, S., Rempfler, M.: Automatic liver and tumor segmentation of CT and MRI volumes using cascaded fully convolutional neural networks. arXiv:1702.05970 (2017)
Zeiler, M.D., Krishnan, D., Taylor, G.W., Fergus, R.: Deconvolutional networks. In: 2010 IEEE Computer Society Conference on computer vision and pattern recognition, pp. 2528–2535 (2010)
Xie, S., Tu, Z.: Holistically-nested edge detection. In: Proceedings of the IEEE international conference on computer vision, pp. 1395–1403 (2015)
Glorot, X., Bengio, Y.: Understanding the difficulty of training deep feedforward neural networks. In: Proceedings of the thirteenth international conference on artificial intelligence and statistics, pp. 249–256 (2010)
Soler, L., Hostettler, A., Agnus, V., Charnoz, A., Fasquel, J., Moreau, J., Marescaux, J.: 3D image reconstruction for comparison of algorithm database: a patient specific anatomical and medical image database. In: IRCAD, Strasbourg, France, Tech. Rep. (2010)
Gauriau, R., Cuingnet, R., Lesage, D., Bloch, I.: Multi-organ localization with cascaded global-to-local regression and shape prior. In: Medical image analysis, pp. 70–83 (2015)
Wolz, R., Chu, C., Misawa, K., Fujiwara, M., Mori, K., Rueckert, D.: Automated abdominal multi-organ segmentation with subject-specific atlas generation. In: IEEE transactions on medical imaging, pp. 1723–1730 (2013)
He, B., Huang, C., Jia, F.: Fully automatic multi-organ segmentation based on multi-boost learning and statistical shape model search. In: VISCERAL Challenge@ ISBI, pp. 18–21 (2015)
Ben-Cohen, A., Diamant, I., Klang, E., Amitai, M., Greenspan, H.: Fully convolutional network for liver segmentation and lesions detection. In: Deep learning and data labeling for medical applications, pp. 77–85 (2016)
Ahmad, M., Yang, J., Ai, D., Qadri, S.F., Wang, Y.: Deep-stacked auto encoder for liver segmentation. In: Chinese conference on image and graphics technologies, pp. 243–251 (2017)
Rafiei, S., Karimi, N., Mirmahboub, B., Soroushmehr, S.M., Felfelian, B., Samavi, S., Najarian, K.: Liver segmentation in abdominal CT images by adaptive 3D region growing. arXiv:1802.07794 (2018)
Acknowledgements
The authors would like to thank Radiologists of the Medical Imaging Department of Affiliated Hospital of Jiangsu University. This work was supported by the National Natural Science Foundation of China (61772242, 62076130, 61572239, 61976106, 91846104), the China Postdoctoral Science Foundation (2017M611737), the Six Talent Peaks Project in Jiangsu Province (DZXX-122) and the Key Special Project of Health and Family Planning Science and Technology in Zhenjiang City (SHW2017019).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Communicated by X. Yang.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Liu, Z., Han, K., Wang, Z. et al. Automatic liver segmentation from abdominal CT volumes using improved convolution neural networks. Multimedia Systems 27, 111–124 (2021). https://doi.org/10.1007/s00530-020-00709-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00530-020-00709-x