Abstract
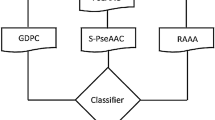
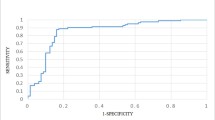
Hormone-binding proteins (HBPs) are important soluble carriers for growth hormones, and correct recognition of HBPs is crucial to understanding their functions. Therefore, we aimed to construct an efficient and reliable classifier to identify HBPs accurately. At first, 246 proteins were collected from UniProt database and considered as the objective benchmark dataset. We employed the 8000-dimensional feature extraction method based on tripeptide compositions to formulate protein samples. Subsequently, we alleviated the intricate feature set by utilizing ANOVA, a feature ranking technique, and acquired the optimal feature subset devoid of redundant information. Furthermore, we utilized three classification methods to process the selected tripeptide features, which generated three probability sequences. Finally, the three probability sequences were considered as new features, and addressed by the support vector machine to construct a prediction model. Results indicated that 90.6% of accuracy was achieved in five-fold cross validation, which was superior to that of other published methods.


Similar content being viewed by others
References
Baumann G (2002) Growth hormone binding protein. The soluble growth hormone receptor. Minerva Endocrinol 27(4):265–276
Dhiraviam KN, Balasubramanian S, Jayavel S (2018) Indole alkaloids as new leads for the design and development of novel DPP-IV inhibitors for the treatment of diabetes. Curr Bioinform 13(2):157–169
Ozzola G (2016) Essay of sex hormone binding protein in internal medicine: a brief review. La Clinica Terapeutica 167(5):e127–e129
Kraut JA, Madias NE (2017) Adverse Effects of the Metabolic Acidosis of Chronic Kidney Disease. Adv Chron Kidney Dis 24(5):289–297
Yang X-G, Luo R-Y, Feng Z-P (2007) Using amino acid and peptide composition to predict membrane protein types. Biochem Biophys Res Commun 353(1):164–169
Liu B et al (2015) Pse-in-one: a web server for generating various modes of pseudo components of DNA, RNA, and protein sequences. Nucle Acids Res 43(W1):W65–W71
Chou KC (2001) Prediction of protein cellular attributes using pseudo-amino acid composition. Proteins Struct Funct Bioinf 43(3):246–255
Liu B et al (2017) Pse-Analysis: a python package for DNA, RNA and protein peptide sequence analysis based on pseudo components and kernel methods. Oncotarget 8(8):13338–13343
Zhang Z-H et al (2006) A novel method for apoptosis protein subcellular localization prediction combining encoding based on grouped weight and support vector machine. FEBS Lett 580(26):6169–6174
Liu B et al (2014) iDNA-Prot|dis: identifying DNA-binding proteins by incorporating amino acid distance-pairs and reduced alphabet profile into the general pseudo amino acid composition. PLoS One 9(9):e106691
Chen Y et al (2003) Secreted protein prediction system combining CJ-SPHMM, TMHMM, and PSORT. Mamm Genome 14(12):859–865
Nakai K, Horton P (1999) PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci 24(1):34–35
Cai Y-D et al (2004) Application of SVM to predict membrane protein types. J Theor Biol 226(4):373–376
Liu H et al (2005) Using Fourier spectrum analysis and pseudo amino acid composition for prediction of membrane protein types. Protein J 24(6):385–389
Liu B et al (2014) Combining evolutionary information extracted from frequency profiles with sequence-based kernels for protein remote homology detection. Bioinformatics 30(4):472–479
Xiao Y, Zhang J, Deng L (2017) Prediction of lncRNA-protein interactions using HeteSim scores based on heterogeneous networks. Sci Rep 7(1):3664
Cai Y-D, Liu X-J, Chou K-C (2001) Artificial neural network model for predicting membrane protein types. J Biomol Struct Dyn 18(4):607–610
Rezaei MA et al (2008) Prediction of membrane protein types by means of wavelet analysis and cascaded neural networks. J Theor Biol 254(4):817–820
Zhang J et al (2018) Ontological function annotation of long non-coding RNAs through hierarchical multi-label classification. Bioinformatics 34(10):1750–1757
Shen H, Chou K-C (2005) Using optimized evidence-theoretic K-nearest neighbor classifier and pseudo-amino acid composition to predict membrane protein types. Biochem Biophys Res Commun 334(1):288–292
Tanchotsrinon W, Lursinsap C, Poovorawan Y (2017) An efficient prediction of HPV genotypes from partial coding sequences by Chaos game representation and fuzzy k-nearest neighbor technique. Curr Bioinform 12(5):431–440
Bulashevska A, Eils R (2006) Predicting protein subcellular locations using hierarchical ensemble of Bayesian classifiers based on Markov chains. BMC Bioinform 7(1):298
Scott MS, Thomas DY, Hallett MT (2004) Predicting subcellular localization via protein motif co-occurrence. Genome Res 14(10a):1957–1966
Deng L, Chen Z (2015) An integrated framework for functional annotation of protein structural domains. IEEE/ACM Trans Comput Biol Bioinform (TCBB) 12(4):902–913
Zeng C, Zhan W, Deng L (2018) SDADB: a functional annotation database of protein structural domains. Database 2018:bay064
Chen Y-L, Li Q-Z (2007) Prediction of the subcellular location of apoptosis proteins. J Theor Biol 245(4):775–783
Chou K-C, Elrod DW (1999) Protein subcellular location prediction. Protein Eng 12(2):107–118
Lin C et al (2013) Hierarchical classification of protein folds using a novel ensemble classifier. PLoS One 8(2):e56499
Song L et al (2014) nDNA-prot: identification of DNA-binding proteins based on unbalanced classification. BMC Bioinform 15(1):298
Zou Q et al (2013) BinMemPredict: a web server and software for predicting membrane protein types. Curr Proteomics 10(1):2–9
Wan S, Duan Y, Zou Q (2017) HPSLPred: an ensemble multi-label classifier for human protein subcellular location prediction with imbalanced source. Proteomics 17:1700262
Zou Q et al (2015) Improving tRNAscan-SE Annotation Results via Ensemble Classifiers. Mol Inform 34(11–12):761–770
Liu B et al (2017) iRSpot-EL: identify recombination spots with an ensemble learning approach. Bioinformatics 33(1):35–41
Liu B et al (2018) iEnhancer-EL: identifying enhancers and their strength with ensemble learning approach. Bioinformatics. https://doi.org/10.1093/bioinformatics/bty458
Liu B, Yang F, Chou K-C (2017) 2L-piRNA: a two-layer ensemble classifier for identifying piwi-interacting RNAs and their function. Mol Ther Nucl 7:267–277
Pan Y, Wang Z, Zhan W, Deng L (2018) Computational identification of binding energy hot spots in protein-RNA complexes using an ensemble approach. Bioinformatics 34(9):1473–1480
Long HX, Wang M, Fu HY (2017) Deep convolutional neural networks for predicting hydroxyproline in proteins. Curr Bioinform 12(3):233–238
Wei L et al (2018) Prediction of human protein subcellular localization using deep learning. J Parallel Distrib Comput 117:212–217
Wei L et al (2019) Integration of deep feature representations and handcrafted features to improve the prediction of N6-methyladenosine sites. Neurocomputing 324:3–9. https://doi.org/10.1016/j.neucom.2018.04.082
Li S, Chen J, Liu B (2017) Protein remote homology detection based on bidirectional long short-term memory. BMC Bioinform 18:443
Tang H et al (2018) HBPred: a tool to identify growth hormone-binding proteins. Int J Biol Sci 14(8):957–964
Niu M et al (2018) RFAmyloid: a web server for predicting amyloid proteins. Int J Mol Sci 19(7):2071
Ding C et al (2012) Identification of mycobacterial membrane proteins and their types using over-represented tripeptide compositions. J Proteomics 77:321–328
Zhu P-P et al (2015) Predicting the subcellular localization of mycobacterial proteins by incorporating the optimal tripeptides into the general form of pseudo amino acid composition. Mol BioSyst 11(2):558–563
Ding H et al (2014) Identification of bacteriophage virion proteins by the ANOVA feature selection and analysis. Mol BioSyst 10(8):2229–2235
Tang H, Chen W, Lin H (2016) Identification of immunoglobulins using Chou’s pseudo amino acid composition with feature selection technique. Mol BioSyst 12(4):1269–1275
Zhao Y-W et al (2017) IonchanPred 2.0: a tool to predict ion channels and their types. Int J Mol Sci 18(9):1838
Holmes G, Donkin A, Witten IH (1994) Weka: A machine learning workbench. In: Proceedings of the 2nd Australian and New Zealand conference on intelligent information systems, 1994. IEEE
Tang H et al (2016) Identification of apolipoprotein using feature selection technique. Sci Rep 6:30441
Liu B et al (2015) Identification of real microRNA precursors with a pseudo structure status composition approach. PLoS One 10(3):e0121501
Chen J et al (2018) A comprehensive review and comparison of different computational methods for protein remote homology detection. Brief Bioinform 9(2):231–244
Wei L et al (2018) ACPred-FL: a sequence-based predictor based on effective feature representation to improve the prediction of anti-cancer peptides. Bioinformatics 34:4007–4016
Fan GL et al (2015) DSPMP: discriminating secretory proteins of malaria parasite by hybridizing different descriptors of C hou’s pseudo amino acid patterns. J Comput Chem 36(31):2317–2327
Gautam A et al (2012) CPPsite: a curated database of cell penetrating peptides. Database 2012
Guo S-H et al (2014) iNuc-PseKNC: a sequence-based predictor for predicting nucleosome positioning in genomes with pseudo k-tuple nucleotide composition. Bioinformatics 30(11):1522–1529
Lin H et al (2014) iPro54-PseKNC: a sequence-based predictor for identifying sigma-54 promoters in prokaryote with pseudo k-tuple nucleotide composition. Nucl Acids Res 42(21):12961–12972
Suratanee A, Plaimas K (2014) Identification of inflammatory bowel disease-related proteins using a reverse k-nearest neighbor search. J Bioinform Comput Biol 12(04):1450017
Wei L, Tang J, Zou Q (2017) Local-DPP: an improved DNA-binding protein prediction method by exploring local evolutionary information. Inf Sci 384:135–144
Zhang J, Liu B (2017) PSFM-DBT: identifying DNA-binding proteins by combing position specific frequency matrix and distance-bigram transformation. Int J Mol Sci 18:1856
Liu Y, Wang X, Liu B (2017) A comprehensive review and comparison of existing computational methods for intrinsically disordered protein and region prediction. Brief Bioinform. https://doi.org/10.1093/bib/bbx126
Su R et al (2018) Developing a multi-dose computational model for drug-induced hepatotoxicity prediction based on toxicogenomics data. IEEE/ACM Trans Comput Biol Bioinform. https://doi.org/10.1109/TCBB.2018.2858756
Liu B et al (2018) iRO-3wPseKNC: identify DNA replication origins by three-window-based PseKNC. Bioinformatics 1:8. https://doi.org/10.1093/bioinformatics/bty312
Tang H et al (2016) Prediction of cell-penetrating peptides with feature selection techniques. Biochem Biophys Res Commun 477(1):150–154
Wei L et al (2018) Prediction of human protein subcellular localization using deep learning. J Parallel Distrib Comput 117:212–217
Wei L et al (2017) Fast prediction of protein methylation sites using a sequence-based feature selection technique. IEEE/ACM Trans Comput Biol Bioinf 1:1
Tang W, Liao Z, Zou Q (2016) Which statistical significance test best detects oncomiRNAs in cancer tissues? An exploratory analysis. Oncotarget 7:85613. https://doi.org/10.18632/oncotarget.12828
Lin C et al (2014) LibD3C: ensemble classifiers with a clustering and dynamic selection strategy. Neurocomputing 123:424–435
Liu B (2017) BioSeq-Analysis: a platform for DNA, RNA and protein sequence analysis based on machine learning approaches. Brief Bioinform. https://doi.org/10.1093/bib/bbx165
Zou Q et al (2014) Survey of MapReduce frame operation in bioinformatics. Brief Bioinform 15(4):637–647
Su W et al (2017) Multiple sequence alignment based on a suffix tree and center-star strategy: a linear method for multiple nucleotide sequence alignment on spark parallel framework. J Comput Biol 24(12):1230–1242
Guo R et al (2018) Bioinformatics applications on Apache Spark. GigaScience 7(8):giy098
Acknowledgements
The work was supported by the National Key R&D Program of China (SQ2018YFC090002), and the Natural Science Foundation of China (Nos. 61771331, 61871282).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, K., Li, S., Wang, Q. et al. Identification of hormone-binding proteins using a novel ensemble classifier. Computing 101, 693–703 (2019). https://doi.org/10.1007/s00607-018-0682-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00607-018-0682-x