Abstract
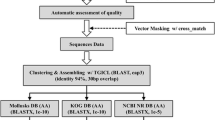
We have developed a new algorithm for invertebrate expressed sequence tag (EST) analysis, termed as the fmEST algorithm, which consists of a systematic homology search, functional motif scanning, and clustering alignment. This study was undertaken to evaluate the validity of our fmEST algorithm in functional motif discovery for invertebrate EST sequence data. Out of 200 unidentified invertebrate ESTs, including 100 arthropod ESTs and 100 mollusk ESTs, 18 arthropod ESTs and 21 mollusk ESTs were identified as fmESTs that contained functional motifs. The nucleotide lengths of arthropod fmEST and mollusk fmEST sequences were distributed from 388 to 954 bp and from 222 to 742 bp, respectively. This result allowed us to annotate these invertebrate fmESTs as various functional genes, while they showed no significant homology to the gene information recorded in the international DNA databases using the conventional BLAST homology search program. In addition, another 1 arthropod EST and 23 mollusk ESTs were assembled into contigs with any identified fmESTs by clustering alignment. Based on these findings, we have concluded that our fmEST algorithm, involving the functional motif discovery procedure, is a valuable approach, enabling us to break new ground in undeveloped invertebrate EST analysis.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.References
E Asamizu Y Nakamura (2004) ArticleTitleSystem for expressed sequence tags (EST) analysis (in Japanese) Tanpakushitsu Kakusan Koso 49 1847–1852
AC Angus ST Ong FT Chew (2004) ArticleTitleSequence tag catalogs of dust-mite-expressed genomes: utility in allergen and acarologic studies Am J Pharmacogenomics 4 357–369 Occurrence Handle10.2165/00129785-200404060-00003
AK Samuels DW Weisrock JJ Smith et al. (2005) ArticleTitleTranscriptional and phylogenetic analysis of five complete ambystomatid salamander mitochondrial genomes Gene 349 43–53 Occurrence Handle10.1016/j.gene.2004.12.037
DL Evans H Kaur J Leary SuffixIII et al. (2005) ArticleTitleMolecular characterization of a novel pattern recognition protein from nonspecific cytotoxic cells Dev Comp Immunol 29 1049–1064 Occurrence Handle10.1016/j.dci.2005.03.011
AC Faria-Campos GC Cerqueira C Anacleto et al. (2003) ArticleTitleMining microorganism EST databases in the quest for new proteins Genet Mol Res 31 169–177
PS Gross TC Bartlett CL Browdy et al. (2001) ArticleTitleImmune gene discovery by expressed sequence tag analysis of hemocytes and hepatopancreas in the Pacific white shrimp, Litopenaeus vannamei, and the Atlantic white shrimp, L. setiferus Dev Comp Immunol 25 565–577 Occurrence Handle10.1016/S0145-305X(01)00018-0
SA Lehnert KJ Wilson K Byrne et al. (1999) ArticleTitleTissue-specific expressed sequence tags from the black tiger shrimp Penaeus monodon Mar Biotechnol 1 465–476 Occurrence Handle10.1007/PL00011803
P Supungul S Klinbunga R Pichyangkura et al. (2004) ArticleTitleAntimicrobial peptides discovered in the black tiger shrimp Penaeus monodon using the EST approach Dis Aquat Organ 61 123–135 Occurrence Handle10.3354/dao061123
MJ Jenny AH Ringwood ER Lacy et al. (2002) ArticleTitlePotential indicators of stress response identified by expressed sequence tag analysis of hemocytes and embryos from the American oyster, Crassostrea virginica Mar Biotechnol 4 81–93 Occurrence Handle10.1007/s10126-001-0072-8
YH Lee GM Huang RA Cameron et al. (1999) ArticleTitleEST analysis of gene expression in early cleavage-stage sea urchin embryos Development 126 3857–3867
EL Ljunggren D Nilsson JG Mattsson (2003) ArticleTitleExpressed sequence tag analysis of Sarcoptes scabiei Parasitology 127 139–145 Occurrence Handle10.1017/S003118200300338X
Ohkubo M, Aranishi F (2004) What does invertebrate EST analysis acquire from bioinformatics? In: Kim J-H (ed) Simulated evolution and learning. KAIST, Daejeon, Korea, Genome Informatics, pp 1–5
JC Whisstock AM Lesk (2003) ArticleTitlePrediction of protein function from protein sequence and structure Q Rev Biophys 36 307–340 Occurrence Handle10.1017/S0033583503003901
JD Thompson DG Higgins TJ Gibson (1994) ArticleTitleCLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice Nucleic Acids Res 22 4673–4680 Occurrence Handle10.1093/nar/22.22.4673
G Pertea X Huang F Liang et al. (2003) ArticleTitleTIGR gene indices clustering tools (TGICL): a software system for fast clustering of large EST datasets Bioinformatics 19 651–652 Occurrence Handle10.1093/bioinformatics/btg034
M Boutros N Perrimon (2000) ArticleTitleDrosophila genome takes flight Nat Cell Biol 2 E53–E54 Occurrence Handle10.1038/35008678
A Hotz-Wagenblatt T Hankeln P Ernst et al. (2003) ArticleTitleEST annotator: a tool for high throughput EST annotation Nucleic Acids Res 31 3716–3719 Occurrence Handle10.1093/nar/gkg566
Author information
Authors and Affiliations
Corresponding author
Additional information
This work was presented in part at the 11th International Symposium on Artificial Life and Robotics, Oita, Japan, January 23–25, 2006
About this article
Cite this article
Ohkubo, M., Aranishi, F. A functional motif discovery algorithm for invertebrate EST sequence data. Artif Life Robotics 11, 135–138 (2007). https://doi.org/10.1007/s10015-006-0415-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10015-006-0415-7