Abstract
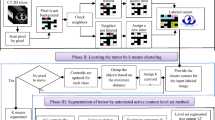
In this article, we propose a lung tumor segmentation algorithm based on the Allen–Cahn (AC) energy equation. The novelty lies in the fact that, when extracting the energy matrix using the AC energy equation, we employ a sliding window algorithm for feature extraction on the data without neglecting local features. After obtaining the energy matrix, we construct constraint conditions based on the minimum and maximum values in the matrix, forming an arithmetic progression. Due to the flexibility in setting the sliding window size and constraint conditions, we can achieve segmentation results according to different requirements. In the numerical experiments, we conduct segmentation experiments of varying difficulty in both two-dimensional (2D) and three-dimensional (3D) spaces to verify the effectiveness of the proposed method. When addressing the lung tumor segmentation problem, we compare the maximum diameter of 3D lung tumors segmented by our proposed segmentation algorithm with the maximum diameter of lung tumors in the original 2D CT images to validate the segmentation accuracy and significance of the proposed method. By conducting more detailed and precise measurements and segmentations of tumors in 3D space, this approach contributes to advancements in medical science and enhances patient treatment outcomes. We also conduct tumor segmentation experiments on the MSD and LIDC-IDRI datasets, setting up comparison metrics to further verify the method’s effectiveness.














Similar content being viewed by others
Data availability
Publicly available data are used.
References
Hussain SP, Hofseth LJ, Harris CC (2003) Radical causes of cancer. Nat Rev Cancer 3(4):276–285. https://doi.org/10.1038/nrc1046
Cardenas CE, Yang J, Anderson BM et al (2019) Advances in auto-segmentation. Semin Radiat Oncol 29(3):185–197. https://doi.org/10.1016/j.semradonc.2019.02.001
Pezzano G, Ripoll VR, Radeva P (2021) CoLe-CNN: Context-learning convolutional neural network with adaptive loss function for lung nodule segmentation. Comput Meth Prog Bio 198:105792. https://doi.org/10.1016/j.cmpb.2020.105792
Zhang B, Qi S, Wu Y et al (2022) Multi-scale segmentation squeeze-and-excitation UNet with conditional random field for segmenting lung tumor from CT images. Comput Meth Prog Bio 222:106946. https://doi.org/10.1016/j.cmpb.2022.106946
Primakov SP, Ibrahim A, van Timmeren JE et al (2022) Automated detection and segmentation of non-small cell lung cancer computed tomography images. Nat Commun 13(1):3423. https://doi.org/10.1038/s41467-022-30841-3
Li N, Tan F, Chen W et al (2022) One-off low-dose CT for lung cancer screening in China: a multicentre, population-based, prospective cohort study. Lancet Respir Med 10(4):378–391. https://doi.org/10.1016/S2213-2600(21)00560-9
Yang J, Veeraraghavan H, van Elmpt W et al (2020) CT images with expert manual contours of thoracic cancer for benchmarking auto?segmentation accuracy. Med Phys 47(7):3250–3255. https://doi.org/10.1002/mp.14107
Shi J, Ye Y, Zhu D et al (2021) Comparative analysis of pulmonary nodules segmentation using multiscale residual U-Net and fuzzy C-means clustering. Comput Meth Prog Bio 209:106332. https://doi.org/10.1016/j.cmpb.2021.106332
Divya S, Padma Suresh L, John A (2022) Enhanced deep-joint segmentation with deep learning networks of glioma tumor for multi-grade classification using MR images. Pattern Anal Appl 25(4):891–911. https://doi.org/10.1007/s10044-022-01064-5
Navaneethakrishnan M, Anand MV, Vasavi G (2023) Deep Fuzzy SegNet-based lung nodule segmentation and optimized deep learning for lung cancer detection. Pattern Anal Appl 8:1–17. https://doi.org/10.1007/s10044-023-01135-1
Qu S, Wang Z, Wu J (2024) FBRNet: a feature fusion and border refinement network for real-time semantic segmentation. Pattern Anal Appl 8:1–18. https://doi.org/10.1007/s10044-023-01207-2
Bouchot A, Ferrieux-Paquet A, Mollon G, Descartes S, Debayle J (2022) Segmentation and morphological analysis of wear track/particles images using machine learning. J Electron Imaging 31(5):051605–051605. https://doi.org/10.1117/1.JEI.31.5.051605
Nugroho A, Hidayat R, Adi Nugroho H, Debayle J (2020) Cancerous object detection using morphological region-based active contour in ultrasound images. J Phys Conf Ser 2:12011. https://doi.org/10.1088/1742-6596/1444/1/012011
Dutande P, Baid U, Talbar S (2022) Deep residual separable convolutional neural network for lung tumor segmentation. Comout Biol Med 141:105161. https://doi.org/10.1016/j.compbiomed.2021.105161
Feng Y, Hafiane A, Laurent H (2024) A weakly supervised end-to-end framework for semantic segmentation of cancerous area in whole slide image. Pttern Anal Appl 27(2):35. https://doi.org/10.1007/s10044-024-01251-6
Gan W, Wang H, Gu H et al (2021) Automatic segmentation of lung tumors on CT images based on a 2D & 3D hybrid convolutional neural network. Brit J Radiol 94:20210038. https://doi.org/10.1259/bjr.20210038
Yang J, Wu B, Li L et al (2021) MSDS-UNet: a multi-scale deeply supervised 3D U-Net for automatic segmentation of lung tumor in CT. Comput Med Imag Grap 92:101957. https://doi.org/10.1016/j.compmedimag.2021.101957
Pei X, Ren Y, Tang Y, Wang Y, Zhang L, Wei J, Zhao D (2024) MFDiff: multiscale feature diffusion model for segmentation of 3D intracranial aneurysm from CT images. Pttern Anal Appl 27(2):66. https://doi.org/10.1007/s10044-024-01266-z
Chen W, Yang F, Zhang X, Xu X, Qiao X (2021) MAU-Net: multiple attention 3D U-Net for lung cancer segmentation on CT images. Proc Comput Sci 192:543–552. https://doi.org/10.1016/j.procs.2021.08.056
Zhang R, Cheng C, Zhao X, Li X (2019) Multiscale mask R-CNN-based lung tumor detection using PET imaging. Mol Imaging 18:1536012119863531. https://doi.org/10.1177/1536012119863531
Zhou J, Kuang H, Wang Y, Wang J (2024) Combining CNN and self-attention-free transformer using local-global attention fusion for lung cancer segmentation. In: International Conference on Intelligent Computing, pp. 371-380 https://doi.org/10.1007/978-981-97-5692-6_33
Shakeel PM, Burhanuddin MA, Desa MI (2022) Automatic lung cancer detection from CT image using improved deep neural network and ensemble classifier. Neural Comput Appl 2:1–14. https://doi.org/10.1007/s00521-020-04842-6
Wang M, Li D (2022) An automatic segmentation method for lung tumor based on improved region growing algorithm. Diagnostics 12(12):2971. https://doi.org/10.3390/diagnostics12122971
Wang S, Mahon R, Weiss E et al (2023) Automated lung cancer segmentation using a PET and CT dual-modality deep learning neural network. Int J Radiat Oncol 115(2):529–539. https://doi.org/10.1016/j.ijrobp.2022.07.2312
Li M, Ma Y, Huang H (2023) Expanded relative density peak clustering for image segmentation. Pattern Anal Appl 26(4):1685–1701. https://doi.org/10.1007/s10044-023-01195-3
Huang X, Wang Q, Chen J et al (2023) Effective hybrid attention network based on pseudo-color enhancement in ultrasound image segmentation. Image Vision Comput 104:742. https://doi.org/10.1016/j.imavis.2023.104742
Chen LQ, Zhao Y (2022) From classical thermodynamics to phase-field method. Prog Mater Sci 124:100868. https://doi.org/10.1016/j.pmatsci.2021.100868
Zhuang X, Zhou S, Huynh GD et al (2022) Phase field modeling and computer implementation: a review. Eng Fract Mech 262:108234. https://doi.org/10.1016/j.engfracmech.2022.108234
Cui C, Ma R, Martínez-Pañeda E (2021) A phase field formulation for dissolution-driven stress corrosion cracking. J Mech Phys Solids 147:104254. https://doi.org/10.1016/j.jmps.2020.104254
Lo YS, Hughes TJ, Landis CM (2023) Phase-field fracture modeling for large structures. J Mech Phys Solids 171:105118. https://doi.org/10.1016/j.jmps.2022.105118
Abaza A, Laurencin J, Nakajo A, Meille S, Debayle J, Leguillon D (2022) Prediction of crack nucleation and propagation in porous ceramics using the phase-field approach. Theor Appl Fract Mec 119:103349. https://doi.org/10.1016/j.tafmec.2022.103349
Allen SM, Cahn JW (1979) A microscopic theory for antiphase boundary motion and its application to antiphase domain coarsening. Acta Metall 27(6):1085–1095. https://doi.org/10.1016/0001-6160(79)90196-2
Beneš M, Chalupecký V, Mikula K (2004) Geometrical image segmentation by the Allen-Cahn equation. Appl Numer Math 51(2–3):187–205. https://doi.org/10.1016/j.apnum.2004.05.001
Ham S, Kim JS (2023) Stability analysis for a maximum principle preserving explicit scheme of the Allen-Cahn equation. Math Comput Simul 207:453–465. https://doi.org/10.1016/j.matcom.2023.01.016
Wang J, Han Z, Jiang W et al (2023) A novel classification method combining Phase-Field and DNN. Pattern Recogn 109:723. https://doi.org/10.1016/j.patcog.2023.109723
Wang J, Han Z, Jiang W et al (2023) A fast, efficient, and explicit phase-field model for 3D mesh denoising. Appl Math Comput 458:128239. https://doi.org/10.1016/j.amc.2023.128239
Xia B, Yu R, Song X, Zhang X, Kim JS (2023) An efficient data assimilation algorithm using the Allen–Cahn equation. Eng Anal Bound Elem 155:511–517. https://doi.org/10.1016/j.enganabound.2023.06.029
Wang J, Lee C, Lee HG et al (2021) Phase-field modeling and numerical simulation for ice melting. Numer Math-Theory Me 14(2):540–558. https://doi.org/10.4208/nmtma.OA-2020-0023
Han Z, Xu H, Wang J (2023) A simple shape transformation method based on phase-field model. Comput Math Appl 147:121–129. https://doi.org/10.1016/j.camwa.2023.07.020
Zhou X, Hayashi T, Hara T et al (2006) Automatic segmentation and recognition of anatomical lung structures from high-resolution chest CT images. Comput Med Imag Grap 30(5):299–313. https://doi.org/10.1016/j.compmedimag.2006.06.002
Mansoor A, Bagci U, Foster B et al (2015) Segmentation and image analysis of abnormal lungs at CT: current approaches, challenges, and future trends. Radiographics 35(4):1056–1076. https://doi.org/10.1148/rg.2015140232
De Mello EVL, da Silveira Filho OT (2005) Numerical study of the Cahn-Hilliard equation in one, two and three dimensions. Phys A 347:429–443. https://doi.org/10.1016/j.physa.2004.08.076
Wang J, Han Z, Jiang W, Kim JS (2023) A fast, efficient, and explicit phase-field model for 3D mesh denoising. Appl Math Comput 458:128239. https://doi.org/10.1016/j.amc.2023.128239
Simpson AL, Antonelli M, Bakas S, et al (2019) A large annotated medical image dataset for the development and evaluation of segmentation algorithms. arXiv preprint arXiv:2019. 1902.09063 https://doi.org/10.48550/arXiv.1902.09063
Armato SG III, McLennan G, Bidaut L et al (2015) Data from LIDC-IDRI [Data set]. The Cancer Imaging Archive. https://doi.org/10.7937/K9/TCIA.2015.LO9QL9SX
Isensee F, Petersen J, Klein A, Zimmerer D, Jaeger PF, Kohl S, Maier-Hein K H (2018) nnu-net: Self-adapting framework for u-net-based medical image segmentation. arXiv preprint arXiv: 2018. 1809.10486 https://doi.org/10.48550/arXiv.1809.10486
Yu Q, Yang D, Roth H, Bai Y, Zhang Y, Yuille AL, Xu D (2020) C2fnas: Coarse-to-fine neural architecture search for 3d medical image segmentation. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 4126-4135 https://doi.org/10.48550/arXiv.1912.09628
Zhou Y, Li Z, Bai S, Wang C, Chen X, Han M, Yuille AL (2019) Prior-aware neural network for partially-supervised multi-organ segmentation. In: Proceedings of the IEEE/CVF international conference on computer vision, pp. 10672-10681 https://doi.org/10.48550/arXiv.1904.06346
Liu W, Qi Y, Li J, Ren Z (2023) Lung nodule segmentation based on complementary context-aware networks. In: 2023 42nd Chinese Control Conference (CCC), pp. 7705-7710 https://doi.org/10.23919/CCC58697.2023.10240177
Wu B, Zhou Z, Wang J, Wang Y (2018) Joint learning for pulmonary nodule segmentation, attributes and malignancy prediction. In: 15th IEEE International Symposium on Biomedical Imaging, pp. 1109-1113 https://doi.org/10.1109/ISBI.2018.8363765
Wang S et al (2017) A multi-view deep convolutional neural networks for lung nodule segmentation. In: 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, pp. 1752-1755 https://doi.org/10.1109/EMBC.2017.8037182
Wang S et al (2017) Central focused convolutional neural networks: developing a data-driven model for lung nodule segmentation. Med Image Anal 40:172–183. https://doi.org/10.1016/j.media.2017.06.014
Mukherjee S, Huang X, Bhagalia RR (2017) Lung nodule segmentation using deep learned prior based graph cut. In: 14th IEEE International Symposium on Biomedical Imaging, pp. 1205-1208 https://doi.org/10.1109/ISBI.2017.7950733
Nithila EE, Kumar SS (2016) Segmentation of lung nodule in CT data using active contour model and Fuzzy C-mean clustering. Alex Eng J 55(3):2583–2588. https://doi.org/10.1016/j.aej.2016.06.002
Hernández-Solis V, Téllez-Velázquez A, Orantes-Molina A, Cruz-Barbosa R (2021) Lung-nodule segmentation using a convolutional neural network with the U-net architecture. In: Mexican Conference on Pattern Recognition, pp. 335-344 https://doi.org/10.1007/978-3-030-77004-4_32
Acknowledgements
The authors express gratitude to the reviewers for their constructive feedback and suggestions, which have significantly enhanced the quality of this paper.
Funding
The first author (Jian Wang) expresses thanks for the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (Grant Nos. 22KJB110020) and the support by the Open Project of Center for Applied Mathematics of Jiangsu Province (Nanjing University of Information Science and Technology). The corresponding author (J.S. Kim) was supported by the National Research Foundation(NRF), Korea, under project BK21 FOUR.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no Conflict of interest regarding the publication of this article.
Ethics approval
Not applicable. No data were collected within the scope of this work. All data sets used have been collected and made available by other authors. Therefore, no ethical approval is required.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, J., Han, Z., Chen, X. et al. A fast and accurate 3D lung tumor segmentation algorithm. Pattern Anal Applic 28, 42 (2025). https://doi.org/10.1007/s10044-025-01425-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10044-025-01425-w