Abstract
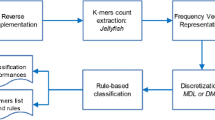
Biological macromolecules, i.e. DNA, RNA and proteins, are coded by strings, called primary structures. During the last decades, the number and the complexity of primary structures are growing exponentially. Analyzing this huge volume of data to extract pertinent knowledge is a challenging task. Data mining approaches can be helpful to reach this goal. In this paper, we present a new data mining approach, called Disclass, based on vote strategies to do classification of primary structures: Let f1,f2,...,f n be families that represent, respectively, n samples of n sets S1,S2,...,S n of primary structures. Let us consider now a new primary structure w that is assumed to belong to one of the n sets S1,S2,...,S n . By using our data mining approach Disclass, the decision to assign the new primary structure w to one of the sets S1,S2,...,S n is taken as follows: (i) During the first step, for each family f i , 1≤i≤n, we construct the ambiguously discriminant and minimal substrings (ADMS) associated with this family. Because the family f i , 1≤i≤n, is a sample of the set S i , the obtained ADMS are considered also to be associated with the whole set S i . During the classification process, the ADMS associated with the set S i , that are approximate substrings of the new primary structure w, will vote with weighted voices for the set S i . (ii) During the second step, we compute according to a vote strategy, the voice weights of the different ADMS, constructed during the first step. (iii) Finally, during the last step, the set that has the maximum weight of voices is the set to which we assign the new primary structure w.
Similar content being viewed by others
References
Beyer WA, Stein ML, Smith TF, Ulman SM (1974) A molecular-sequence metric and evolutionary tree. Math Biosci 19:9–25
Burge Ch (1997) Identification of genes in human genomic DNA. Dissertation, Stanford University, Stanford, CA
Craven MW, Shavlik JW (1994) Machine learning approaches to gene recognition. IEEE Expert 9(2):2–10
Day WHE, Johnson DS, Sankoff D (1986) The computational complexity of inferring rooted phylogenies by parsimony. Math Biosci 81:33–42
Dayhoff MO, Schwartz RM, Orcutt BC (1978) A model for evolutionary change. Atlas Prot Seq Struct 5(3):345–352
Doolittle RF (1990) Molecular evolution: computer analysis of protein and nucleic acid sequences. Meth Enzymol 183
Doolittle RF (ed) (1990) Molecular evolution: computer analysis of protein and nucleic acid sequences. Meth Enzymol 183
Elloumi M (1994) Analysis of strings coding biological macromolecules. Dissertation, The University of Aix-Marseilles III, France
Elloumi M (1998) Comparison of strings belonging to the same family. Inf Sci Int J 111(1–4):49–63
Elloumi M (2001) An algorithm for the approximate string-matching problem. In: Proceedings of Atlantic symposium on computational biology, genome information systems & technology, Durham, NC
Fu H (2001) A study of amino acids binary codes. Dissertation, University of Lille, France
Gusfield D (1990) Efficient algorithms for inferring evolutionary trees. Networks 21:19–28
Henikoff S, Henikoff JG (1992) Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA 89:10915–10919
Hirsh H, Noordewier M (1994) Using background knowledge to improve inductive learning of DNA sequences. In: Proceedings of the tenth IEEE conference on artificial intelligence for applications, San Antonio, TX, pp 351–357
Hirsh JD, Sternberg MJE (1992) Prediction of structural and functional features of protein and nucleic acid sequences by artificial neural networks. Biochemistry 31(32):7211–7218
Kannan S, Warnow T (1990) Inferring evolutionary history from DNA sequences. In: Proceedings of 31st annual IEEE symposium on foundation of computer science, St. Louis, MO, pp 326–371
Karp RM, Miller RE, Rosenberg AL (1972) Rapid identification of repeated patterns in strings, trees and arrays. In: Proceedings of the fourth annual ACM symposium on theory of computing, Denver, CO, pp 125–136
Krogh A, Brown M, Mian IS, Sjolander K, Haussler D (1994) Hidden Markov models in computational biology: applications to protein modeling. J Mol Biol 235(5):1501–1531
Levenshtein VI (1966) Binary codes capable of correcting deletions, insertions and reversals. Cybernet Control Theory 10(8):707–710
Maddouri M (2000) Contribution to concept learning: towards an incremental approach to induce production rules from examples. Dissertation, Faculty of Sciences of Tunis, The University of Tunis-El Manar
Maddouri M, Elloumi M (2002) A data mining approach based on machine learning techniques to classify biological sequences. Knowl Based Syst Jl 15(4):217–223
Mironov AA, Roytberg MA, Pevzner PA, Gelfand MS (1998) Performance-guarantee gene predictions via spliced sequence alignment. Genomics 51:332–339
O’Neill MC (1989) Consensus methods for finding and ranking DNA binding sites. J Mol Biol 207:301–310
O’Neill MC, Chiafari F (1989) Escherichia coli promoters. II: a spacing class-dependent promoter search protocol. J Biol Chem 264:5531–5534
Opitz DW, Shavlik JW (1997) Connectionist theory refinement: genetically searching the space of network topologies. J Artif Intell Res 6:177–209
Qicheng M, Wang JTL, Gattiker JR (2002) Mining biomolecular data using background knowledge and artificial neural networks. Research Report, Dept of Computer and Information Science, New Jersey Institute of Technology, New Jersey
Quinlan JR (1983) Learning efficient classification procedures and their application to chess end-games. In: Michalski RS, Carbonell JG, Mitchell TM (eds) Machine learning: an artificial intelligence approach. Tioga, Palo Alto, CA, pp 463–482
Salzberg S, Delcher A, Heath D, Kasif S (1995) Best-case results for nearest-neighbor learning. IEEE Trans Pattern Anal Mach Intell 17(6):599–608
Schiffmann W, Joost M, Werner R (1994) Optimization of the backpropagation algorithm for training multilayer perceptrons. Technical Report, Institute of Physics, Rheinau 1, University of Koblenz, Koblenz
Sze SH, Roytberg MA, Gelfand MS, Mironov AA, Astakhova TV, Pevzner PA (1998) Algorithms and software for support of gene identification experiments. Bioinformatics 14(1):14–19
Towell GG (1991) Symbolic knowledge and neural networks: insertion, refinement and extraction. Dissertation, Dept of Computer Sciences, University of Wisconsin–Madison
Towell GG, Shavlik JW, Noordenier MO (1990) Refinement of approximate domain theories by knowledge-based artificial neural networks. In: Proceedings of the Eighth national conference on artificial intelligence, Boston, MA, pp 861–866
Wang JTL, Marr TG, Shasha D, Shapiro B, Chirn GW (1994) Discovering active motifs in sets of related protein sequences and using them for classification. Nucleic Acids Res 22:2769–2775
Wang JTL, Ma Q, Shasha D, Wu CH (2001) New techniques for extracting features from protein sequences. IBM Syst Jl 40(2):426–441
Weiss SM, Kulikowski CA (1991) Computer systems that learn. Kaufmann, California
Zurada JM (1992) Introduction to artificial neural systems. PWS, Boston, MA, pp 186–196
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Elloumi, M., Maddouri, M. New voting strategies designed for the classification of nucleic sequences. Knowl Inf Syst 8, 1–15 (2005). https://doi.org/10.1007/s10115-004-0151-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10115-004-0151-z