Abstract
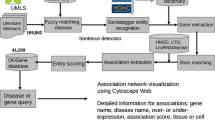
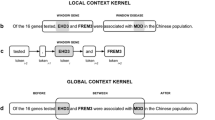
Objective Advanced experimental methods such as next-generation sequencing (NGS) produced a large number of potential indicative genetic biomarkers and gene variants to diseases mentioned as research outputs in the scientific literature. To elucidate novel biomarkers and therapeutic candidates from this larger number of literature, highly sophisticated text mining-based knowledge-driven frameworks are a necessity. Materials and Methods This paper presents DisGeReExT Web server for performing a literature-wide analysis study (LWAS) to extract both direct and indirect gene–disease associations using joint ensemble learning (explicit) along with concept profiling using the ABC principle (implicit) for prioritizing and rationalizing potential informative discoveries of the genetic role on diseases. In addition, we ranked the informative sentences using a scoring model and calculated the disease–disease similarity using functional association among shared genes. Results From complete MEDLINE corpus dated September 2020 with 28 million records, DisGeReExT identified a total of 2,237,545 gene–disease associations and 2,851,662 disease–disease similarities. Discussion DisGeReExT was able to extract informative sentences related to both diseases and genes in large scale. It also explored the gene–disease association of two diseases, namely Alzheimer’s disease and liver carcinoma, and identified its top 10 associated genes and diseases of both diseases. Conclusion Overall, we strongly believe that our large-scale automated approach for knowledge discovery of gene-associated diseases from literature could provide new insights into the genetic mechanism and disease etiology and can play a pivotal role in translational research, drug discovery, and repurposing.








Similar content being viewed by others
Data availability
DisGeReExT is freely available at http://14.139.186.184/DisGeReExT/.
Notes
DisGeReExT is freely available at http://14.139.186.184/DisGeReExT/.
References
Menche J, Sharma A, Kitsak M et al (2015) Uncovering disease-disease relationships through the incomplete interactome. Science. https://doi.org/10.1126/science.1257601
Rifai N, Gillette MA, Carr SA (2006) Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotechnol. https://doi.org/10.1038/nbt1235
Aerts S, Lambrechts D, Maity S et al (2006) Gene prioritization through genomic data fusion. Nat Biotechnol. https://doi.org/10.1038/nbt1203
Timpson NJ, Greenwood CMT, Soranzo N et al (2018) Genetic architecture: the shape of the genetic contribution to human traits and disease. Nat Rev Genet. https://doi.org/10.1038/nrg.2017.101
Nielsen J (2017) Systems biology of metabolism: a driver for developing personalized and precision medicine. Cell Metab. https://doi.org/10.1016/j.cmet.2017.02.002
Seyyedrazzagi E, Navimipour NJ (2017) Disease genes prioritizing mechanisms: a comprehensive and systematic literature review. Netw Model Anal Heal Inform Bioinform. https://doi.org/10.1007/s13721-017-0154-9
Hamazaki T, El Rouby N, Fredette NC et al (2017) Concise review: Induced pluripotent stem cell research in the era of precision medicine. Stem Cells. https://doi.org/10.1002/stem.2570
Peng J, Bai K, Shang X et al (2017) Predicting disease-related genes using integrated biomedical networks. BMC Genom. https://doi.org/10.1186/s12864-016-3263-4
Goh K, Cusick ME, Valle D et al (2007) The human disease network. Proc Natl Acad Sci USA. https://doi.org/10.1073/pnas.0701361104
Lee J, Yoon W, Kim S et al (2020) BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. https://doi.org/10.1093/bioinformatics/btz682
Singhal A, Simmons M, Lu Z (2016) Text mining for precision medicine: automating disease-mutation relationship extraction from biomedical literature. J Am Med Inform Assoc. https://doi.org/10.1093/jamia/ocw041
Pyysalo S, Baker S, Ali I et al (2019) LION LBD: a literature-based discovery system for cancer biology. Bioinformatics. https://doi.org/10.1093/bioinformatics/bty845
Jung JY, DeLuca TF, Nelson TH et al (2014) A literature search tool for intelligent extraction of disease-associated genes. J Am Med Informatics Assoc. https://doi.org/10.1136/amiajnl-2012-001563
Bhasuran B, Subramanian D, Natarajan J (2018) Text mining and network analysis to find functional associations of genes in high altitude diseases. Comput Biol. https://doi.org/10.1016/j.compbiolchem.2018.05.002
Bhasuran B, Natarajan J (2019) Distant supervision for large-scale extraction of gene–disease associations from literature using DeepDive. In: Lecture notes in networks and systems. https://doi.org/10.1007/978-981-13-2354-6_39
Zhao S, Su C, Lu Z et al (2020) Recent advances in biomedical literature mining. Brief Bioinform. https://doi.org/10.1093/bib/bbaa057
Amberger JS, Bocchini CA, Schiettecatte F et al (2015) OMIM.org: online mendelian inheritance in man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. https://doi.org/10.1093/nar/gku1205
Safran M, Dalah I, Alexander J et al (2010) GeneCards version 3: the human gene integrator. Database (Oxford). https://doi.org/10.1093/database/baq020
Pletscher-Frankild S, Pallejà A, Tsafou K et al (2015) DISEASES: text mining and data integration of disease-gene associations. Methods. https://doi.org/10.1016/j.ymeth.2014.11.020
Liu Y, Liang Y, Wishart D (2015) PolySearch2: a significantly improved text-mining system for discovering associations between human diseases, genes, drugs, metabolites, toxins and more. Nucleic Acids Res. https://doi.org/10.1093/nar/gkv383
Piñero J, Ramírez-Anguita JM, Saüch-Pitarch J et al (2020) The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. https://doi.org/10.1093/nar/gkz1021
Kim J, So S, Lee HJ et al (2013) DigSee: disease gene search engine with evidence sentences (version cancer). Nucleic Acids Res. https://doi.org/10.1093/nar/gkt531
Song M, Kim WC, Lee D et al (2015) PKDE4J: entity and relation extraction for public knowledge discovery. J Biomed Inform. https://doi.org/10.1016/j.jbi.2015.08.008
Xu D, Zhang M, Xie Y et al (2016) DTMiner: identification of potential disease targets through biomedical literature mining. Bioinformatics. https://doi.org/10.1093/bioinformatics/btw503
Bhasuran B, Natarajan J (2018) Automatic extraction of gene-disease associations from literature using joint ensemble learning. PLoS One. https://doi.org/10.1371/journal.pone.0200699
Tsuruoka Y, Tsujii J, Ananiadou S (2008) FACTA: a text search engine for finding associated biomedical concepts. Bioinformatics. https://doi.org/10.1093/bioinformatics/btn469
Tsuruoka Y, Miwa M, Hamamoto K et al (2011) Discovering and visualizing indirect associations between biomedical concepts. Bioinformatics. https://doi.org/10.1093/bioinformatics/btr214
Jelier R, Jenster G, Dorssers LCJ et al (2007) Text-derived concept profiles support assessment of DNA microarray data for acute myeloid leukemia and for androgen receptor stimulation. BMC Bioinformatics. https://doi.org/10.1186/1471-2105-8-14
Jelier R, Schuemie MJ, Veldhoven A et al (2008) Anni 2.0: a multipurpose text-mining tool for the life sciences. Genome Biol. https://doi.org/10.1186/gb-2008-9-6-r96
Cheng D, Knox C, Young N et al (2008) PolySearch: a web-based text mining system for extracting relationships between human diseases, genes, mutations, drugs and metabolites. Nucleic Acids Res. https://doi.org/10.1093/nar/gkn296
Fleuren WWM, Verhoeven S, Frijters R et al (2011) CoPub update: CoPub 5.0 a text mining system to answer biological questions. Nucleic Acids Res. https://doi.org/10.1093/nar/gkr310
ElShal S, Tranchevent LC, Sifrim A et al (2016) Beegle: from literature mining to disease-gene discovery. Nucleic Acids Res. https://doi.org/10.1093/nar/gkv905
Tranchevent LC, Barriot R, Yu S et al (2008) ENDEAVOUR update: a web resource for gene prioritization in multiple species. Nucleic Acids. https://doi.org/10.1093/nar/gkn325
Kim J, Kim JJ, Lee H (2017) An analysis of disease-gene relationship from medline abstracts by DigSee. Sci Rep. https://doi.org/10.1038/srep40154
Hettne KM, Thompson M, Van Haagen HHHBM et al (2016) The implicitome: a resource for rationalizing gene-disease associations. PLoS One. https://doi.org/10.1371/journal.pone.0149621
Fontaine JF, Andrade-Navarro MA (2016) Gene set to diseases (GS2D): disease enrichment analysis on human gene sets with literature data. Genomics Comput Biol. https://doi.org/10.18547/gcb.2016.vol2.iss1.e33
Swanson DR 1991 Complementary structures in disjoint science literatures. In: Proceedings of the 14th annual international ACM SIGIR conference on research and development in information retrieval, SIGIR 1991. https://doi.org/10.1145/122860.122889
Crichton G, Baker S, Guo Y et al (2020) Neural networks for open and closed literature-based discovery. PLoS One. https://doi.org/10.1371/journal.pone.0232891
Xie Q, Yang KM, Heo GE et al (2020) Literature based discovery of alternative TCM medicine for adverse reactions to depression drugs. BMC Bioinformatics. https://doi.org/10.1186/s12859-020-03735-8
Weeber M, Vos R, Klein H et al (2003) Generating hypotheses by discovering implicit associations in the literature: a case report of a search for new potential therapeutic uses for thalidomide. J Am Med Inform Assoc. https://doi.org/10.1197/jamia.M1158
Weeber M, Klein H et al (2001) Using concepts in literature-based discovery: simulating Swanson’s Raynaud-fish oil and migraine-magnesium discoveries. J Am Soc Inf Sci Technol. https://doi.org/10.1002/asi.1104b
Swanson DR (1986) Fish oil, Raynaud’s syndrome, and undiscovered public knowledge. Perspect Biol Med. https://doi.org/10.1353/pbm.1986.0087
Digiacomo RA, Kremer JM, Shah DM (1989) Fish-oil dietary supplementation in patients with Raynaud’s phenomenon: a double-blind, controlled, prospective study. Am J Med. https://doi.org/10.1016/0002-9343(89)90261-1
Gopalakrishnan V, Jha K, Jin W et al (2019) A survey on literature based discovery approaches in biomedical domain. J Biomed Inform. https://doi.org/10.1016/j.jbi.2019.103141
Kim YH, Song M (2019) A context-based ABC model for literature-based discovery. PLoS One. https://doi.org/10.1371/journal.pone.0215313
Thilakaratne M, Falkner K, Atapattu T (2019) A systematic review on literature-based discovery workflow. PeerJComput Sci. https://doi.org/10.7717/peerj-cs.235
Bhasuran B, Murugesan G, Abdulkadhar S et al (2016) Stacked ensemble combined with fuzzy matching for biomedical named entity recognition of diseases. J Biomed Inform. https://doi.org/10.1016/j.jbi.2016.09.009
Murugesan G, Abdulkadhar S, Bhasuran B et al (2017) BCC-NER: bidirectional, contextual clues named entity tagger for gene/protein mention recognition. Eurasip J Bioinforma Syst Biol. https://doi.org/10.1186/s13637-017-0060-6
Bodenreider O (2004) The unified medical language system (UMLS): integrating biomedical terminology. Nucleic Acids Res. https://doi.org/10.1093/nar/gkh061
Lipscomb CE (2000) Medical subject headings (MeSH). Bull Med Libr Assoc 88:265
Gray KA, Yates B, Seal RL et al (2015) Genenames.org: the HGNC resources in 2015. Nucleic Acids Res. https://doi.org/10.1093/nar/gku1071
Law V, Knox C, Djoumbou Y et al (2014) DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res. https://doi.org/10.1093/nar/gkt1068
Ursu O, Holmes J, Knockel J et al (2017) DrugCentral: online drug compendium. Nucleic Acids Res. https://doi.org/10.1093/nar/gkw993
Mao Y, Lu Z (2017) MeSH now: automatic MeSH indexing at PubMed scale via learning to rank. J Biomed Semantics. https://doi.org/10.1186/s13326-017-0123-3
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. https://doi.org/10.1111/j.2517-6161.1995.tb02031.x
Ferro N, Crestani F, Moens MF, et al. Beyond factoid QA: effective methods for non-factoid answer sentence retrieval. Lect Notes Comput Sci (including SubserLect Notes ArtifIntellLect Notes Bioinformatics) 2016
Ferreira R, De Souza CL, Lins RD et al (2013) Assessing sentence scoring techniques for extractive text summarization. Expert Syst Appl. https://doi.org/10.1016/j.eswa.2013.04.023
Dang V (2012) The lemur project-wiki-ranklib. Lemur Project
Wei CH, Harris BR, Kao HY et al (2013) TmVar: A text mining approach for extracting sequence variants in biomedical literature. Bioinformatics. https://doi.org/10.1093/bioinformatics/btt156
Demeester T, Sutskever I, Chen K, et al. (2016) Distributed representations of words and phrases and their compositionality. EMNLP 2016-Conf Empir Methods Nat Lang Process Proc
Ibrahim OAS, Landa-Silva D (2018) An evolutionary strategy with machine learning for learning to rank in information retrieval. Soft Comput. https://doi.org/10.1007/s00500-017-2988-6
You Y, Lu C, Wang W et al (2019) Relative CNN-RNN: learning relative atmospheric visibility from images. IEEE Trans Image Process. https://doi.org/10.1109/TIP.2018.2857219
Wu Q, Burges CJC, Svore KM et al (2010) Adapting boosting for information retrieval measures. Inf Retr Boston. https://doi.org/10.1007/s10791-009-9112-1
Real R, Vargas JM (1996) The probabilistic basis of Jaccard’s index of similarity. Syst Biol. https://doi.org/10.1093/sysbio/45.3.380
Hoehndorf R, Schofield PN, Gkoutos GV (2015) Analysis of the human diseasome using phenotype similarity between common, genetic, and infectious diseases. Sci Rep. https://doi.org/10.1038/srep10888
Rayner L, McGovern A, Creagh-Brown B et al (2018) Type 2 diabetes and asthma: systematic review of the bidirectional relationship. Curr Diabetes Rev. https://doi.org/10.2174/1573399814666180711114859
Masoudkabir F, Sarrafzadegan N, Gotay C et al (2017) Cardiovascular disease and cancer: Evidence for shared disease pathways and pharmacologic prevention. Atherosclerosis. https://doi.org/10.1016/j.atherosclerosis.2017.06.001
Žitnik M, Janjić V, Larminie C et al (2013) Discovering disease-disease associations by fusing systems-level molecular data. Sci Rep. https://doi.org/10.1038/srep03202
Cheng L, Jiang Y, Wang Z et al (2016) DisSim: an online system for exploring significant similar diseases and exhibiting potential therapeutic drugs. Sci Rep. https://doi.org/10.1038/srep30024
Suthram S, Dudley JT, Chiang AP et al (2010) Network-based elucidation of human disease similarities reveals common functional modules enriched for pluripotent drug targets. PLoSComput Biol. https://doi.org/10.1371/journal.pcbi.1000662
Mathur S, Dinakarpandian D (2012) Finding disease similarity based on implicit semantic similarity. J Biomed Inform. https://doi.org/10.1016/j.jbi.2011.11.017
Davis AP, Rosenstein MC, Wiegers TC et al (2011) DiseaseComps: a metric that discovers similar diseases based upon common toxicogenomic profiles at CTD. Bioinformation. https://doi.org/10.6026/97320630007154
Davis AP, Wiegers TC, King BL et al (2016) Generating gene ontology-disease inferences to explore mechanisms of human disease at the comparative toxicogenomics database. PLoS One. https://doi.org/10.1371/journal.pone.0155530
Gligorijevic D, Stojanovic J, Djuric N et al (2016) Large-scale discovery of disease-disease and disease-gene associations. Sci Rep. https://doi.org/10.1038/srep32404
Acknowledgements
This work was supported by DRDO—BU Centre for Life Sciences, Coimbatore, Tamil Nadu, India. BB acknowledges the fellowship received from the grant.
Author information
Authors and Affiliations
Contributions
All authors have made a substantial, direct, and intellectual contribution to this study.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bhasuran, B., Natarajan, J. DisGeReExT: a knowledge discovery system for exploration of disease–gene associations through large-scale literature-wide analysis study. Knowl Inf Syst 65, 3463–3487 (2023). https://doi.org/10.1007/s10115-023-01862-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10115-023-01862-1