Abstract
Although ultrasound plays an important role in the diagnosis of chronic kidney disease (CKD), image interpretation requires extensive training. High operator variability and limited image quality control of ultrasound images have made the application of computer-aided diagnosis (CAD) challenging. This study assessed the effect of integrating computer-extracted measurable features with the convolutional neural network (CNN) on the ultrasound image CAD accuracy of CKD. Ultrasound images from patients who visited Severance Hospital and Gangnam Severance Hospital in South Korea between 2011 and 2018 were used. A Mask regional CNN model was used for organ segmentation and measurable feature extraction. Data on kidney length and kidney-to-liver echogenicity ratio were extracted. The ResNet18 model classified kidney ultrasound images into CKD and non-CKD. Experiments were conducted with and without the input of the measurable feature data. The performance of each model was evaluated using the area under the receiver operating characteristic curve (AUROC). A total of 909 patients (mean age, 51.4 ± 19.3 years; 414 [49.5%] men and 495 [54.5%] women) were included in the study. The average AUROC from the model trained using ultrasound images achieved a level of 0.81. Image training with the integration of automatically extracted kidney length and echogenicity features revealed an improved average AUROC of 0.88. This value further increased to 0.91 when the clinical information of underlying diabetes was also included in the model trained with CNN and measurable features. The automated step-wise machine learning–aided model segmented, measured, and classified the kidney ultrasound images with high performance. The integration of computer-extracted measurable features into the machine learning model may improve CKD classification.
Keywords: Chronic kidney disease, Computer-aided diagnosis, Convolutional neural network, Deep learning, Kidney ultrasound, Machine learning
Introduction
Differentiating chronic kidney disease (CKD) from acute kidney injury (AKI) is the most fundamental step in the management of impaired kidney function. AKI refers to a sudden loss of kidney function, whereas in CKD, kidney damage usually persists for some amount of time. When the cause of kidney damage is properly identified and removed, kidney function eventually recovers in AKI [1]. However, in patients with CKD, kidney damage is irreversible and often progressive, leading to end-stage renal disease (ESRD) [2]. Periodic blood test results that reflect kidney function are useful in distinguishing CKD from AKI. However, when such data are not obtainable, making an accurate diagnosis of CKD versus AKI is often challenging.
Imaging studies are key diagnostic modalities for identifying CKD [3, 4]. In particular, ultrasound imaging is the most important scheme used to detect CKD in clinical practice [5–7]. The well-known advantages of ultrasound imaging, including its low cost and noninvasive nature and lack of ionizing radiation, have made it an attractive option for detecting kidney diseases [8]. The characteristic changes in kidney length, echogenicity, and corticomedullary differentiation enable a CKD diagnosis without the need for invasive procedures such as kidney biopsy [9]. However, kidney ultrasound image acquisition and interpretation are prone to operator dependency [10–12]. Ultrasound-based diagnosis is highly dependent on the ability and experience of the person performing ultrasonography, and therefore requires extensive training and experience for optimal use. This limits its widespread application in facilities or regions lacking well-experienced specialists.
The application of machine learning in medical image analysis has been widely studied [13, 14]. Machine learning algorithms are helpful for reducing false-positive results in computed tomography (CT) image lung nodule detection [15, 16]. In addition, the ability to identify the malignant potential of breast nodules on CT and magnetic resonance imaging (MRI) images was greatly improved with machine learning–based computer-aided diagnosis (CAD) [17, 18]. Accumulating studies have also attempted to apply machine learning techniques to improve the detection rates of ultrasound imaging [19, 20]. However, machine learning–obtained diagnostic accuracy in ultrasound was often inferior to that performed by experienced radiologist [21, 22]. This would be due to the ultrasound-specific characteristics such as low image quality and high inter- and intra-observer variability [10].
In this study, assessments were performed to evaluate whether integrating data on measurable features into a convolutional neural network (CNN) could improve the CAD accuracy of CKD in ultrasound imaging. Segmentation of the kidney and extraction of measurable features, such as kidney length and echogenicity, were performed using deep learning methods.
Material and Methods
Study Design and Subjects
Patients who underwent kidney ultrasound imaging between 2011 and 2018 at Severance Hospital and Gangnam Severance Hospital in South Korea were screened. Patients with estimated glomerular filtration rate (eGFR) ≥ 90 or ≤ 60 were initially selected for inclusion to compare imaging of patients with normal eGFR and patients with CKD [23, 24]. Patients who met the following criteria were excluded: (1) age < 18 years; (2) ESRD with dialysis at the time of ultrasound imaging; (3) previous kidney transplantation; (4) previous nephrectomy; (5) known kidney cysts, solid mass, or hydronephrosis; and (6) missing liver or kidney images on ultrasound. A total of 909 patients were included in the final analysis. This study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of Yonsei University Health System Clinical Trial Center (1–2018-0039). The need for informed consent was waived by the institutional review board owing to the study’s retrospective design.
Patient Classification
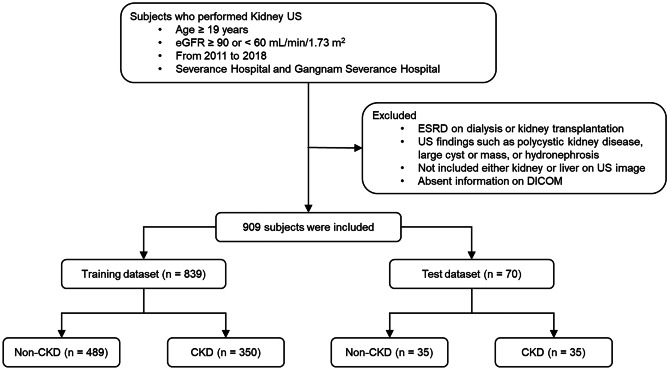
Serum creatinine levels were determined using an isotope dilution mass spectrometry–traceable method at a central laboratory. The eGFR was calculated using the Chronic Kidney Disease Epidemiology Collaboration (CKD-EPI) creatinine equation [25]. Laboratory results within 3 months before kidney ultrasound imaging were used for patient classification. Those whose eGFR was ≥ 90 mL/min/1.73 m2 within the prior 3 months were labeled the non-CKD group. Those whose eGFR was < 60 mL/min/1.73 m2 throughout the prior 3 months were classified as the CKD group (Fig. 1).
Fig. 1.
Flow diagram of the included participants
Data Collection and De-Identification
Demographic and laboratory data were retrieved from electronic medical records. Data on the presence of comorbidities at the time of ultrasound imaging, including hypertension, diabetes, and coronary artery disease, were also collected. All kidney ultrasound images were obtained by board-certified radiologists. Ultrasound machines (Philips Healthcare, [Amsterdam, Netherlands], Siemens Healthineers [Erlangen, Germany], and GE Healthcare [Chicago, Illinois, USA]) were used for the ultrasound image acquisition. For training and testing, ultrasound images containing both kidney and liver were used. The number of ultrasound images including both the kidney and liver differed for each patient. An average of 3.7 images were employed for evaluation. Although the number of ultrasound images varied for each patient, a single prediction for each patient was derived using average voting. Therefore, the weights of patients were equally used for evaluation. The ultrasound images were transferred as Digital Imaging and Communications in Medicine (DICOM) files, and pixel data were converted to portable network graphics files. Images that did not include annotations for segmentation or length measurements were selected for the analysis. In addition, patients’ personal information was deleted from the image files before the evaluation. This de-identification program was newly developed for the study. It blacked out the upper corners of the images with personal data to mask the identifying information. Since this program only modified the upper corners of the images, it did not fabricate the ultrasound images of the organs that were used for analysis.
Kidney and Liver Segmentation
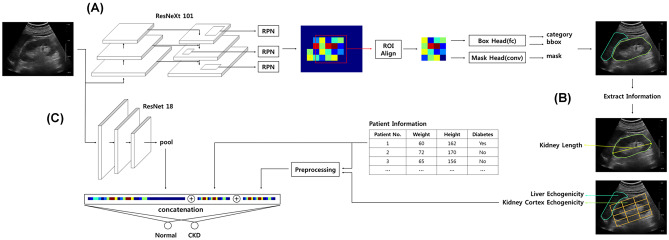
A Mask regional convolutional neural network (R-CNN) model, which adds an additional head to the Faster R-CNN model, was used for the kidney and liver segmentation [26, 27]. The Mask R-CNN is commonly used for instance segmentation where each pixel is assigned an object class and instance identity. This instance segmentation method performs segmentation in a two-step manner: initial detection of the object region followed by object segmentation within the detected area. The U-Net and fully convolutional network, which is widely used in semantic segmentation techniques in medical image segmentation, requires post-processing to filter out small segments and remove holes within the segmentation [28–30]. However, the instance segmentation method requires no additional processing for incorrect segmentation removal. In addition, fewer holes were created compared to one-step methods. The Mask R-CNN models used ImageNet-pretrained ResNeXt101 as the backbone [26, 31]. For feature extraction, ROIAlign and Feature Pyramid Networks (FPN) were used [32]. The ultrasound images were input into the backbone, and the inference and recombined features were extracted from each layer through the FPN. The extracted features were resampled at different scales and ratios at each anchor using ROIAlign and inputted into the Mask R-CNN head for detection, classification, and segmentation. Multi-task loss was used during training. Softmax cross-entropy for classification, smooth L1 for bounding box regression, and binary cross-entropy were used for segmentation. To obtain the trained segmentation model, the combined loss was minimized using a gradient descent (Fig. 2A).
Fig. 2.
The schematic diagram of the algorithm used. A Mask R-CNN model was used to perform segmentation of the kidney and liver (A) and extract measurable features (B). ResNet18 model was used to classify the kidney ultrasound images into CKD or non-CKD (C). CKD, chronic kidney disease; R-CNN, regional convolutional neural network; ROI, region of interest; RPN, region proposal network
Segmentation, Training, and Validation
For segmentation training and validation, 9407 ultrasound images were acquired from 456 patients. A trained technician manually depicted the two-dimension regions of the kidney and liver under nephrologist supervision. A total of 3917 kidney segmentations and 7234 liver segmentations were collected (some images contained both kidney and liver); of them, 993 images were randomly selected for validation. The stochastic gradient descent was utilized with a batch size of 48, and the learning rate started at 0.001 and decreased by 0.1 at 30,000 steps and 40,000 steps. The model was trained in 50,000 steps. The segmentation performance was validated on the validation dataset every 2500 steps using dice coefficient, and the most accurate model was selected.
Extraction of Measurable Features
Measurable features were extracted from the segmented ultrasound images using the Mask R-CNN model. The kidney length was derived by calculating the longest horizontal length of the segmented kidney. To derive the actual length (cm) of the kidney, the kidney length in the image space was calibrated using the machine information stored in the DICOM file. Measuring the absolute level of echogenicity is difficult since echogenicity varies with ultrasound machine operating settings and patient’s state of hydration. Therefore, kidney cortical echogenicity is generally graded relative to the liver parenchyma [33]. Liver echogenicity was obtained by averaging the pixels of the segmented liver area. For calculation of the kidney cortical echogenicity, the segmented kidney was first divided into 3 × 4 grid cells along the longest horizontal axis. The top middle cell was considered to represent cortical echogenicity, and the pixels in this area were averaged (Fig. 2B).
Classification of Kidney Ultrasound Image
For the classification model, the ResNet18 model was used [34]. This model classified kidney ultrasound images into CKD or non-CKD. To input meaningful areas in the classification model, the kidney region was cropped into a rectangular shape aligned with the longest horizontal axis of the kidney segmentation. ResNet uses a skip connection to the convolutional layer, where the input features are added to convoluted features. After the convolutional stage was finished, global average pooling was applied to the convoluted output feature, and a fully connected layer was used to classify the images as CKD or non-CKD. For classifications considering both the image feature and the extracted measurable feature, the features were pre-processed via standardization (mean of 0 and standard deviation of 1) and combined through channel-wise concatenation to the penultimate layer and input to the following final fully connected layer. Softmax cross-entropy was used to calculate the error between the true label and the model prediction (Fig. 2C).
Classification, Training, and Testing
Using the model trained in the segmentation, a total of 3495 ultrasound images that contained both the kidney and the liver were acquired from 909 patients. These images were used for the classification training and testing. For the training dataset, 839 patients (3001 images, 92%) were randomly selected. The remaining 70 patients (247 images, 8%) were used as the testing dataset. For the validation dataset, the patients in the training dataset were divided into 10 groups. Consequently, 10 validation datasets were constructed by selecting one group for validation (approximately 249 images [8%]) and the others for training. To reduce the difference in brightness between ultrasound manufacturers, images that contained both kidney and liver segments were selected, and the kidney echogenicity was normalized against that of the liver. The adaptive moment estimation optimizer was used with a batch size of 16 and a learning rate of 0.001, and the learning rate was reduced to 90 epochs following the cosine annealing learning rate scheduler [35, 36]. In addition, the augmentation techniques (random flip, translate, rotate, scale, contrast, noise, dropout, and blur) were used for training. To ensure reliability, 10 training sessions were conducted on one validation dataset (tenfold cross-validation), resulting in a total of 100 training processes (10 training sessions with different training seeds for each of the 10 different validation datasets). For each training process, the most accurate model was chosen, and the best model was evaluated in the test dataset. Three types of experiments were conducted: “image training without any measurable features,” “image training with extracted measurable features,” and “image training with extracted measurable features and diabetes information.” Each of the three experiments was trained with a randomly initialized model and an ImageNet-pretrained model, resulting in a total of 600 experiments. The final classification results were then calculated for each subject. In cases where multiple ultrasound images included both kidney and liver per subject, each network probability output was averaged to yield one diagnostic result. These evaluations were performed on Intel i7-8700 K and Nvidia GeForce GTX 1080 Ti.
Statistical Methods
Variables were expressed as mean and standard deviation (SD) for continuous variables and frequency and percentage (%) for categorical variables. Intergroup comparisons were performed using Student’s t-test for continuous variables and Pearson’s chi-squared test for categorical variables. The CNN-based CKD classification was compared with the diagnosis of CKD based on eGFR, while the model performance was evaluated using the area under the receiver operating characteristic curve (AUROC). All statistical analyses were performed using SPSS Statistics v.23 (IBM Corporation, Armonk, NY, USA) and STATA v.15.1 (Stata Corporation, College Station, TX, USA). Statistical significance was set at two-sided values of p < 0.05.
Results
Baseline Characteristics
The mean participant age was 51.4 ± 19.3 years; 414 (49.5%) of them were male (Table 1). The non-CKD group comprised 524 (57.6%) participants, while 385 (42.4%) participants were allocated to the CKD group. The CKD group was older and had a higher prevalence of hypertension, diabetes, and coronary artery disease. The average eGFR was 115.5 ± 29.1 and 27.3 ± 11.0 mL/min/1.73 m2 in the non-CKD and CKD groups, respectively.
Table 1.
Patients’ baseline characteristics
| Total (n = 909) | Normal (n = 524) | CKD (n = 385) | P | |
|---|---|---|---|---|
| Age, years | 51.4 ± 19.3 | 42.4 ± 17.6 | 63.7 ± 14.1 | < 0.001 |
| Male sex, n (%) | 414 (45.5) | 212 (40.5) | 202 (52.5) | < 0.001 |
| Height, cm | 162.9 ± 8.9 | 163.6 ± 9.0 | 161.9 ± 8.6 | < 0.004 |
| Weight, kg | 62.8 ± 14.2 | 63.0 ± 15.8 | 62.6 ± 11.6 | < 0.71 |
| Hypertension, n (%) | 476 (52.4) | 161 (30.7) | 315 (81.8) | < 0.001 |
| Diabetes, n (%) | 231 (25.4) | 71 (13.5) | 160 (41.6) | < 0.001 |
| CAD, n (%) | 134 (14.7) | 44 (8.4) | 90 (23.4) | < 0.001 |
| eGFR, mL/min/1.73 m2 | 78.2 ± 49.4 | 115.5 ± 29.1 | 27.3 ± 11.0 | < 0.001 |
Values are expressed as mean ± SD for continuous variables and frequency (%) for categorical variables
CAD coronary artery disease, CKD chronic kidney disease, eGFR estimated glomerular filtration rate
Kidney Segmentation
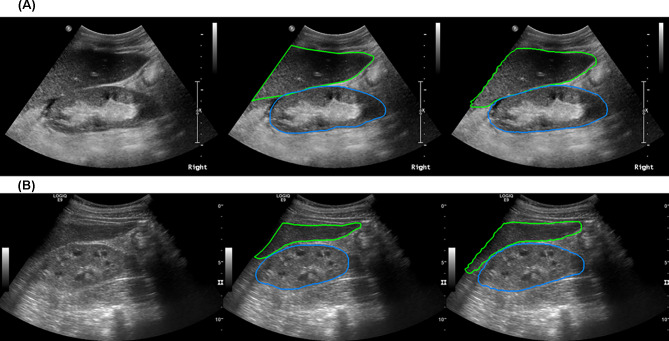
To evaluate the performance of the segmentation implemented by the Mask R-CNN model, its segmented area was compared to the area depicted by trained technicians. In the validation dataset, the best model achieved a mean intersection on union (mIoU) of 81.62, while the frequency-weighted intersection on union (fwIoU) was 89.35 (Fig. 3A and B).
Fig. 3.
Kidney segmentation using Mask R-CNN model. The best model achieved an mIoU of 81.62 and an fwIoU of 89.35. A and B are representative images of kidney and liver segmentation that were achieved by the Mask R-CNN model. fwIoU, frequency-weighted intersection on union; mIoU, mean intersection on union; R-CNN, regional convolutional neural network
Measurable Feature Extraction
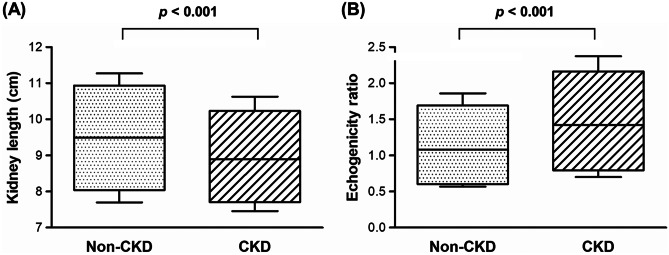
Measurable features, including kidney length and cortical echogenicity, were also obtained using the Mask R-CNN model. The average kidney length was 9.49 ± 1.11 cm in the non-CKD group and 8.90 ± 0.94 cm in the CKD group (p < 0.001; Fig. 4A). When the kidney-to-liver echogenicity ratio was compared between the non-CKD and CKD groups, the echogenicity was significantly higher in the CKD group than in the non-CKD group (1.08 ± 0.44 vs. 1.42 ± 0.53, p < 0.001; Fig. 4B).
Fig. 4.
Extracted measurable features by the Mask R-CNN model. A The average lengths of the kidney were 9.49 ± 1.11 cm and 8.90 ± 0.94 cm in non-CKD and CKD groups, respectively. B The ratios of echogenicity of the kidney to liver were 1.08 ± 0.44 and 1.42 ± 0.53 in non-CKD and CKD groups, respectively. CKD, chronic kidney disease; R-CNN, regional convolutional neural network
CKD Detection Performance Using Ultrasound Image Information Only
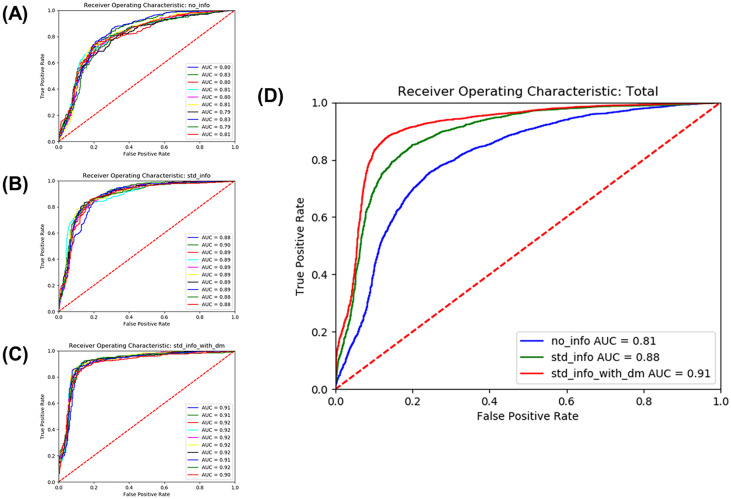
The model performance was evaluated using a test dataset that had been excluded from the training beforehand. Three experiments were completed, and 10 training sessions were performed for each of the 10 validation set experiments. The average AUROC from the model trained with ultrasound images achieved only a level of 0.81 (sensitivity 78.2%, specificity 71.5%, and accuracy 74.4%) (Fig. 5).
Fig. 5.
ROC curve of CKD detection performance. Three types of experiments were conducted. There were 10 validation datasets for each experiment, and the AUROC was obtained for each dataset (A–C). The average AUROC was 0.81 in training without any measurable features, 0.88 in image training with extracted measurable features, and 0.91 in image training with extracted measurable features and diabetes information, respectively (D). AUROC, area under the receiver operating characteristic curve; ROC, receiver operating characteristic
CKD Detection Performance After Integrating Deep Learning–Extracted Measurable Features
The performance of CKD detection from the model trained with ultrasound images only was compared with models that integrated deep learning–extracted measurable feature data and information on diabetes. When data on measurable features, including kidney length and kidney-to-liver echogenicity ratio, were extracted through deep learning algorithms, the average AUROC for CKD detection improved to 0.88 (sensitivity 86.1%, specificity 77.5%, and accuracy 81.2%). Adding information on the presence of diabetes among the participants into the training models resulted in a CKD detection AUROC of 0.91 (sensitivity 89.4%, specificity 82.9%, and accuracy 85.9%) (Fig. 5).
Discussion
In this study, CKD was detected using kidney ultrasound image analysis using deep learning methods. A sequential step-by-step process was employed for the image analyses. First, kidney and liver segmentations were performed using a Mask R-CNN model. Next, the measurable features were extracted from the segmented kidney images. Features such as kidney length and relative cortical echogenicity compared to that of the liver were extracted from the images. Concomitantly, the kidney images were analyzed using the ResNet18 model for CKD detection. Finally, three different models, the image-only model, the model including extracted measurement data, and the model also including diabetes history information, were used for CKD detection. Through this systematic process, the accuracy of detecting CKD was significantly increased when computer-extracted measurable features were added to the machine learning models.
Recently, machine learning algorithms have been applied to increase the accuracy and practicality of medical image analyses. [15–18] Using CNN methods, the detection of breast cancer in screening mammograms revealed an AUC of 0.98 [37]. Functional magnetic resonance imaging (fMRI) data were analyzed using LeNet-5 to detect Alzheimer’s disease, which resulted in a mean accuracy of 96.86% [38]. Although employing machine learning algorithms to medical image data has resulted in high accuracy, the number of systems utilizing ultrasound images is limited compared to CT or MRI [39, 40]. Several investigations have achieved relatively high accuracy, such as the study that applied a three-layer deep belief network in time-intensive curves extracted from contrast-enhanced US (CEUS) video sequences to classify malignant and benign focal liver lesions, which revealed an accuracy of 86.36% [41]. However, most of the attempts at applying machine learning to ultrasound images have resulted in lower accuracies than other imaging modalities. In this study, the AUROC for CKD classification was significantly improved by the imputing of measurable feature data that were automatically extracted through a machine learning process in addition to the CNN-based analysis. This improvement in accuracy shows that imputing supplementary data that could be obtained from the images themselves may compensate for the low accuracy of the CNN-based methods for evaluating ultrasound images.
The CKD-detecting machine learning process developed in this study features an end-to-end characteristic. Each step of segmentation of the organs, extraction of measurable features, and CNN-based classification proceeds automatically without the need for manual intervention. Machine learning methods were assigned to each of these steps. An overall CKD classification accuracy of 85.6% was recently obtained by training kidney ultrasound images through a ResNet model [42]. However, in that study, the boundaries of the kidneys were cropped based on kidney length annotations that were manually performed by sonographers. Although the accuracy of CKD classification (85.9%) was comparable, it should be noted that a fully automated process from segmentation to classification was applied in the current investigation. Compared to previous machine learning–based classification systems, this end-to-end automated workflow would be a noteworthy advantage for practical application in the clinical field.
The accessibility to medical ultrasound imaging is relatively higher than that of other imaging modalities, such as CT and MRI. In addition to the relatively lower cost associated with equipment installation, ultrasound machines require much less space and are portable [6, 7]. These characteristics allow ultrasound to be the primary choice for organ imaging in developing countries and in regions that have poor medical access. However, the comparatively low image quality and inter-operator variability were the main limitations of ultrasound, which has prevented it from being more widely used [10]. In this regard, the presented automated machine learning–based CKD classification model may help overcome the shortcomings of ultrasound diagnosis. Considering the recent increase in CKD incidence in developing countries, the clinical usefulness of automated high-precision models is high. In addition, the strategy of combining measurable data with CNN to improve ultrasound classification accuracy could also be applied to diagnose other diseases of various organs. Since high-prevalence diseases such as liver diseases [41] and breast [43] and thyroid cancer [44] are initially screened through ultrasound imaging, methods for improving machine learning–based classification accuracy are imperative.
This study has several limitations. First, kidney ultrasound images with large cysts, solid masses, and hydronephrosis were not included in the analysis. The decision to exclude these disease states was made so that a working model solely focused on CKD classification could be developed. However, real-world ultrasound images would include various findings, and a system capable of differentiating the concomitant abnormalities would be needed for actual clinical use. Second, in order to compare the images of normal eGFR patients with those with chronic kidney disease, images from patients whose eGFRs were between 60 and 90 were not collected. eGFR higher than 90 is considered normal kidney function, while the presence of chronic kidney disease is suspected when eGFR is lower than 60 [23, 24]. This selection criterion would affect the real-world efficiency of the developed system. Nonetheless, this conceptual study shows that the integration of computer-extracted measurable features improves the classification accuracy of image machine learning models. Third, although a fully automated process was applied to classify the images for CKD detection, the ultrasound images were initially acquired by clinicians. This would have resulted in inter-operator variability, which could have introduced bias to the analysis. Forth, the fact that the validation sets used for segmentation and classification were not identical could result in overfitting the training set on the segmentation task. This may lead to an overestimation of overall performance. In addition, since the pixel normalization for values of kidney to liver was performed per dataset rather than per-image basis, a possibility of data leak which may have resulted in performance augmentation should also be considered. Finally, the ultrasound images were not standardized because of the study’s retrospective nature. Ultrasound images were uploaded at various angles, and the ultrasound machines used also varied. However, the variability of ultrasound machines may enable view-invariant properties, which may increase the practicality of real-world applications.
Conclusion
The automated step-wise machine learning–aided model successfully segmented the kidney and liver on ultrasound images. It was also effective at extracting measurable features, such as kidney size and echogenicity. The integration of computer-extracted measurable features into the machine learning model resulted in a significantly improved CKD classification. Further investigations are needed to apply this model in the real world.
Author Contribution
S. L. and J. T. P. designed the study; S. L. obtained the patient’s clinical information; Y. A. K. contributed to obtaining ultrasound images; M. K., K. B., and I. H. L. performed machine learning–aided analysis; and S. L. and J. T. P. analyzed the data; S. L. and M. K. made the figures; S. L., M. K., and J. T. P. drafted the manuscript; and S. E. L., I. H. L., S. W. K., and J. T. P. revised the paper. All authors contributed important intellectual content during manuscript drafting or revision, and accepted accountability for the overall work by ensuring that questions pertaining to the accuracy or integrity of any portion of the work were appropriately investigated and resolved. And Sangmi Lee and Myeongkyun Kang contributed equally to this study as co-first authors.
Funding
Not applicable.
Data Availability
Not applicable.
Code Availability
Not applicable.
Declarations
Ethics Approval
This study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of Yonsei University Health System Clinical Trial Center (1–2018-0039).
Consent to Participate
The need for informed consent was waived by the institutional review board owing to the study’s retrospective design.
Conflict of Interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Forni LG, Darmon M, Ostermann M, Oudemans-van Straaten HM, Pettilä V, Prowle JR, Schetz M, Joannidis M. Renal recovery after acute kidney injury. Intensive Care Med. 2017;43(6):855–866. doi: 10.1007/s00134-017-4809-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen TK, Knicely DH, Grams ME. Chronic Kidney Disease Diagnosis and Management: A Review. Jama. 2019;322(13):1294–1304. doi: 10.1001/jama.2019.14745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meola M, Samoni S, Petrucci I. Imaging in Chronic Kidney Disease. Contrib Nephrol. 2016;188:69–80. doi: 10.1159/000445469. [DOI] [PubMed] [Google Scholar]
- 4.Fried JG, Morgan MA. Renal Imaging: Core Curriculum 2019. Am J Kidney Dis. 2019;73(4):552–565. doi: 10.1053/j.ajkd.2018.12.029. [DOI] [PubMed] [Google Scholar]
- 5.Ahmed S, Bughio S, Hassan M, Lal S, Ali M. Role of Ultrasound in the Diagnosis of Chronic Kidney Disease and its Correlation with Serum Creatinine Level. Cureus. 2019;11(3):e4241. doi: 10.7759/cureus.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khati NJ, Hill MC, Kimmel PL. The role of ultrasound in renal insufficiency: the essentials. Ultrasound Q. 2005;21(4):227–244. doi: 10.1097/01.wnq.0000186666.61037.f6. [DOI] [PubMed] [Google Scholar]
- 7.Lucisano G, Comi N, Pelagi E, Cianfrone P, Fuiano L, Fuiano G. Can renal sonography be a reliable diagnostic tool in the assessment of chronic kidney disease? J Ultrasound Med. 2015;34(2):299–306. doi: 10.7863/ultra.34.2.299. [DOI] [PubMed] [Google Scholar]
- 8.Crownover BK, Bepko JL. Appropriate and safe use of diagnostic imaging. Am Fam Physician. 2013;87(7):494–501. [PubMed] [Google Scholar]
- 9.Gulati M, Cheng J, Loo JT, Skalski M, Malhi H, Duddalwar V. Pictorial review: Renal ultrasound. Clin Imaging. 2018;51:133–154. doi: 10.1016/j.clinimag.2018.02.012. [DOI] [PubMed] [Google Scholar]
- 10.Brattain LJ, Telfer BA, Dhyani M, Grajo JR, Samir AE. Machine learning for medical ultrasound: status, methods, and future opportunities. Abdom Radiol (NY) 2018;43(4):786–799. doi: 10.1007/s00261-018-1517-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park SH: Artificial intelligence for ultrasonography: unique opportunities and challenges. Ultrasonography 40(1):3–6, 2021. 10.14366/usg.20078 [DOI] [PMC free article] [PubMed]
- 12.Kim YH: Artificial intelligence in medical ultrasonography: driving on an unpaved road. Ultrasonography 40(3):313–317, 2021. 10.14366/usg.21031 [DOI] [PMC free article] [PubMed]
- 13.Tatsugami F, Higaki T, Nakamura Y, Yu Z, Zhou J, Lu Y, Fujioka C, Kitagawa T, Kihara Y, Iida M, Awai K. Deep learning-based image restoration algorithm for coronary CT angiography. Eur Radiol. 2019;29(10):5322–5329. doi: 10.1007/s00330-019-06183-y. [DOI] [PubMed] [Google Scholar]
- 14.Kolossvary M, De Cecco CN, Feuchtner G, Maurovich-Horvat P. Advanced atherosclerosis imaging by CT: Radiomics, machine learning and deep learning. J Cardiovasc Comput Tomogr. 2019;13(5):274–280. doi: 10.1016/j.jcct.2019.04.007. [DOI] [PubMed] [Google Scholar]
- 15.Song Q, Zhao L, Luo X, Dou X. Using Deep Learning for Classification of Lung Nodules on Computed Tomography Images. J Healthc Eng. 2017;2017:8314740. doi: 10.1155/2017/8314740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Causey JL, Zhang J, Ma S, Jiang B, Qualls JA, Politte DG, Prior F, Zhang S, Huang X. Highly accurate model for prediction of lung nodule malignancy with CT scans. Sci Rep. 2018;8(1):9286. doi: 10.1038/s41598-018-27569-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uhlig J, Uhlig A, Kunze M, Beissbarth T, Fischer U, Lotz J, Wienbeck S. Novel Breast Imaging and Machine Learning: Predicting Breast Lesion Malignancy at Cone-Beam CT Using Machine Learning Techniques. AJR Am J Roentgenol. 2018;211(2):W123–w131. doi: 10.2214/ajr.17.19298. [DOI] [PubMed] [Google Scholar]
- 18.D'Amico NC, Grossi E, Valbusa G, Rigiroli F, Colombo B, Buscema M, Fazzini D, Ali M, Malasevschi A, Cornalba G, Papa S. A machine learning approach for differentiating malignant from benign enhancing foci on breast MRI. Eur Radiol Exp. 2020;4(1):5. doi: 10.1186/s41747-019-0131-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim B, Kim KC, Park Y, Kwon JY, Jang J, Seo JK. Machine-learning-based automatic identification of fetal abdominal circumference from ultrasound images. Physiol Meas. 2018;39(10):105007. doi: 10.1088/1361-6579/aae255. [DOI] [PubMed] [Google Scholar]
- 20.Zheng Q, Furth SL, Tasian GE, Fan Y. Computer-aided diagnosis of congenital abnormalities of the kidney and urinary tract in children based on ultrasound imaging data by integrating texture image features and deep transfer learning image features. J Pediatr Urol. 2019;15(1):75.e71–75.e77. doi: 10.1016/j.jpurol.2018.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chung SR, Baek JH, Lee MK, Ahn Y, Choi YJ, Sung TY, Song DE, Kim TY, Lee JH. Computer-Aided Diagnosis System for the Evaluation of Thyroid Nodules on Ultrasonography: Prospective Non-Inferiority Study according to the Experience Level of Radiologists. Korean J Radiol. 2020;21(3):369–376. doi: 10.3348/kjr.2019.0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Wu Q, Chen Y, Wang Y. A Clinical Assessment of an Ultrasound Computer-Aided Diagnosis System in Differentiating Thyroid Nodules With Radiologists of Different Diagnostic Experience. Front Oncol. 2020;10:557169. doi: 10.3389/fonc.2020.557169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.KDIGO CKD Work Group KDIGO 2012 clinical practice guideline for the evaluation and management of chronic kidney disease. Kidney Int Suppl. 2013;2(1):1–150. doi: 10.1038/ki.2013.243. [DOI] [PubMed] [Google Scholar]
- 24.Inker LA, Astor BC, Fox CH, Isakova T, Lash JP, Peralta CA, Kurella Tamura M, Feldman HI. KDOQI US commentary on the 2012 KDIGO clinical practice guideline for the evaluation and management of CKD. Am J Kidney Dis. 2014;63(5):713–735. doi: 10.1053/j.ajkd.2014.01.416. [DOI] [PubMed] [Google Scholar]
- 25.Du X, Hu B, Jiang L, Wan X, Fan L, Wang F, Cao C. Implication of CKD-EPI equation to estimate glomerular filtration rate in Chinese patients with chronic kidney disease. Ren Fail. 2011;33(9):859–865. doi: 10.3109/0886022x.2011.605533. [DOI] [PubMed] [Google Scholar]
- 26.He K, Gkioxari G, Dollár P, Girshick R, editors: Mask r-cnn. Proceedings of the IEEE international conference on computer vision; 2017
- 27.Ren S, He K, Girshick R, Sun J. Faster r-cnn: Towards real-time object detection with region proposal networks. IEEE transactions on pattern analysis and machine intelligence. 2016;39(6):1137–1149. doi: 10.1109/TPAMI.2016.2577031. [DOI] [PubMed] [Google Scholar]
- 28.Ronneberger O, Fischer P, Brox T, editors: U-net: Convolutional networks for biomedical image segmentation. International Conference on Medical image computing and computer-assisted intervention; 2015: Springer,
- 29.Long J, Shelhamer E, Darrell T, editors: Fully convolutional networks for semantic segmentation. Proceedings of the IEEE conference on computer vision and pattern recognition; 2015 [DOI] [PubMed]
- 30.Golan D, Donner Y, Mansi C, Jaremko J, Ramachandran M: Fully automating Graf’s method for DDH diagnosis using deep convolutional neural networks. Deep Learning and Data Labeling for Medical Applications. Springer, 2016. p. 130–141
- 31.Xie S, Girshick R, Dollár P, Tu Z, He K, editors: Aggregated residual transformations for deep neural networks. Proceedings of the IEEE conference on computer vision and pattern recognition; 2017
- 32.Lin T-Y, Dollár P, Girshick R, He K, Hariharan B, Belongie S, editors: Feature pyramid networks for object detection. Proceedings of the IEEE conference on computer vision and pattern recognition; 2017
- 33.Manley JA, O'Neill WC. How echogenic is echogenic? Quantitative acoustics of the renal cortex. Am J Kidney Dis. 2001;37(4):706–711. doi: 10.1016/s0272-6386(01)80118-9. [DOI] [PubMed] [Google Scholar]
- 34.He K, Zhang X, Ren S, Sun J, editors: Deep residual learning for image recognition. Proceedings of the IEEE conference on computer vision and pattern recognition; 2016
- 35.Kingma DP, Ba J: Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980, 2014.
- 36.Loshchilov I, Hutter F: Sgdr: Stochastic gradient descent with warm restarts. arXiv preprint arXiv:1608.03983, 2016.
- 37.Chougrad H, Zouaki H, Alheyane O: Deep Convolutional Neural Networks for breast cancer screening. Comput Methods Programs Biomed 157:19-30, 2018. 10.1016/j.cmpb.2018.01.011 [DOI] [PubMed]
- 38.Saman Sarraf GT. Classification of Alzheimer’s Disease Using fMRI Data and Deep Learning Convolutional Neural Networks. 2016. arXiv:1603.08631
- 39.Pesapane F, Codari M, Sardanelli F. Artificial intelligence in medical imaging: threat or opportunity? Radiologists again at the forefront of innovation in medicine. Eur Radiol Exp. 2018;2(1):35. doi: 10.1186/s41747-018-0061-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak J, van Ginneken B, Sánchez CI. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 41.Kaizhi Wu XC. Mingyue Ding: Deep learning based classification of focal liver lesions with contrast-enhanced ultrasound. Optik. 2014;125(15):4057–4063. doi: 10.1016/j.ijleo.2014.01.114. [DOI] [Google Scholar]
- 42.Kuo CC, Chang CM, Liu KT, Lin WK, Chiang HY, Chung CW, Ho MR, Sun PR, Yang RL, Chen KT. Automation of the kidney function prediction and classification through ultrasound-based kidney imaging using deep learning. NPJ Digit Med. 2019;2:29. doi: 10.1038/s41746-019-0104-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niell BL, Freer PE, Weinfurtner RJ, Arleo EK, Drukteinis JS. Screening for Breast Cancer. Radiol Clin North Am. 2017;55(6):1145–1162. doi: 10.1016/j.rcl.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 44.Zhou H, Jin Y, Dai L, Zhang M, Qiu Y, Wang K, Tian J, Zheng J. Differential Diagnosis of Benign and Malignant Thyroid Nodules Using Deep Learning Radiomics of Thyroid Ultrasound Images. Eur J Radiol. 2020;127:108992. doi: 10.1016/j.ejrad.2020.108992. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.
Not applicable.