Abstract
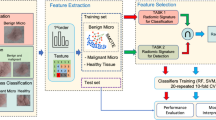
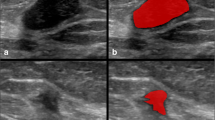
This study investigates the feasibility of using texture radiomics features extracted from mammography images to distinguish between benign and malignant breast lesions and to classify benign lesions into different categories and determine the best machine learning (ML) model to perform the tasks. Six hundred and twenty-two breast lesions from 200 retrospective patient data were segmented and analysed. Three hundred fifty radiomics features were extracted using the Standardized Environment for Radiomics Analysis (SERA) library, one of the radiomics implementations endorsed by the Image Biomarker Standardisation Initiative (IBSI). The radiomics features and selected patient characteristics were used to train selected machine learning models to classify the breast lesions. A fivefold cross-validation was used to evaluate the performance of the ML models and the top 10 most important features were identified. The random forest (RF) ensemble gave the highest accuracy (89.3%) and positive predictive value (66%) and likelihood ratio of 13.5 in categorising benign and malignant lesions. For the classification of benign lesions, the RF model again gave the highest likelihood ratio of 3.4 compared to the other models. Morphological and textural radiomics features were identified as the top 10 most important features from the random forest models. Patient age was also identified as one of the significant features in the RF model. We concluded that machine learning models trained against texture-based radiomics features and patient features give reasonable performance in differentiating benign versus malignant breast lesions. Our study also demonstrated that the radiomics-based machine learning models were able to emulate the visual assessment of mammography lesions, typically used by radiologists, leading to a better understanding of how the machine learning model arrive at their decision.




Similar content being viewed by others
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A: Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer journal for clinicians 68:394-424, 2018
Conti A, Duggento A, Indovina I, Guerrisi M, Toschi N: Radiomics in breast cancer classification and prediction. Semin Cancer Biol, 2020
Crivelli P, Ledda RE, Parascandolo N, Fara A, Soro D, Conti M: A New Challenge for Radiologists: Radiomics in Breast Cancer. Biomed Res Int 2018:6120703, 2018
Castellano G, Bonilha L, Li L, Cendes F: Texture analysis of medical images. Clinical radiology 59:1061-1069, 2004
Zhang Q, et al.: Sonoelastomics for Breast Tumor Classification: A Radiomics Approach with Clustering-Based Feature Selection on Sonoelastography. Ultrasound Med Biol 43:1058-1069, 2017
Tan H, et al.: Mammography-based radiomics nomogram: A potential biomarker to predict axillary lymph node metastasis in breast cancer. The British Journal of Radiology 93:20191019, 2020
Dasgupta A, et al.: Quantitative ultrasound radiomics using texture derivatives in prediction of treatment response to neo-adjuvant chemotherapy for locally advanced breast cancer. Oncotarget 11:3782, 2020
Bickelhaupt S, et al.: Radiomics Based on Adapted Diffusion Kurtosis Imaging Helps to Clarify Most Mammographic Findings Suspicious for Cancer. Radiology 287:761-770, 2018
Gillies RJ, Schabath MB: Radiomics Improves Cancer Screening and Early Detection. Cancer Epidemiol Biomarkers Prev 29:2556-2567, 2020
Schindelin J, et al.: Fiji: an open-source platform for biological-image analysis. Nature methods 9:676-682, 2012
Ashrafinia S: Quantitative nuclear medicine imaging using advanced image reconstruction and radiomics. Johns Hopkins University, 2019
Breiman L: Random forests. Machine learning 45:5-32, 2001
Pal M: Random forest classifier for remote sensing classification. International journal of remote sensing 26:217-222, 2005
Ali J, Khan R, Ahmad N, Maqsood I: Random forests and decision trees. International Journal of Computer Science Issues (IJCSI) 9:272, 2012
Liaw A, Wiener M: Classification and regression by randomForest. R news 2:18-22, 2002
Chen S, et al.: A New Application of Multimodality Radiomics Improves Diagnostic Accuracy of Nonpalpable Breast Lesions in Patients with Microcalcifications-Only in Mammography. Med Sci Monit 25:9786-9793, 2019
Mao N, et al.: Added Value of Radiomics on Mammography for Breast Cancer Diagnosis: A Feasibility Study. J Am Coll Radiol 16:485-491, 2019
Funding
This study was approved by the Medical Ethics Committee of the University of Malaya Medical Centre in 2019 (MECID: 2019822–7771) and supported by the Malaysian Ministry of Higher Education, Fundamental Research Grant Scheme (FRGS) [FRGS/1/2019/SKK03/UM/01/1].
Author information
Authors and Affiliations
Contributions
Study concepts and design came from J H D Wong and L K Tan. The funding source was secured by K Rahmat. Literature research was done by N Letchumanan. Imaging and data were collected and curated by W Y Chan. Image pre-processing and segmentation were performed by N A Mumin. Software codes for texture extraction and machine learning (ML) models were developed by N Letchumanan and L K Tan. Texture analysis and performance of ML models were carried out by N Letchumanan under the supervision of J H D Wong and T K Tan. The manuscript was prepared by L K Tan, J H D Wong, N Letchumanan, and N A Mumin. All authors contributed to editing the manuscript. L K Tan, J H D Wong, and N Letchumanan revised and validate the manuscript. The final version of the manuscript was read and approved by all authors.
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Letchumanan, N., Wong, J.H.D., Tan, L.K. et al. A Radiomics Study: Classification of Breast Lesions by Textural Features from Mammography Images. J Digit Imaging 36, 1533–1540 (2023). https://doi.org/10.1007/s10278-022-00753-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-022-00753-1