Abstract
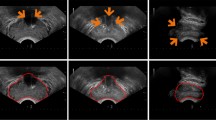
Accurate prostate segmentation in ultrasound images is crucial for the clinical diagnosis of prostate cancer and for performing image-guided prostate surgery. However, it is challenging to accurately segment the prostate in ultrasound images due to their low signal-to-noise ratio, the low contrast between the prostate and neighboring tissues, and the diffuse or invisible boundaries of the prostate. In this paper, we develop a novel hybrid method for segmentation of the prostate in ultrasound images that generates accurate contours of the prostate from a range of datasets. Our method involves three key steps: (1) application of a principal curve-based method to obtain a data sequence comprising data coordinates and their corresponding projection index; (2) use of the projection index as training input for a fractional-order-based neural network that increases the accuracy of results; and (3) generation of a smooth mathematical map (expressed via the parameters of the neural network) that affords a smooth prostate boundary, which represents the output of the neural network (i.e., optimized vertices) and matches the ground truth contour. Experimental evaluation of our method and several other state-of-the-art segmentation methods on datasets of prostate ultrasound images generated at multiple institutions demonstrated that our method exhibited the best capability. Furthermore, our method is robust as it can be applied to segment prostate ultrasound images obtained at multiple institutions based on various evaluation metrics.










Similar content being viewed by others
Data Availability
Data will be made available on reasonable request.
References
D. Karimi, Q. Zeng, P. Mathur, A. Avinash, S. Mahdavi, I. Spadinger, P. Abolmaesumi, S.E. Salcudean, Accurate and robust deep learning-based segmentation of the prostate clinical target volume in ultrasound images, Med. Image Anal. 57 (2019) 186–196.
M.A. Kollmeier, Combined brachytherapy and ultra-hypofractionated radiotherapy for intermediate-risk prostate cancer: Comparison of toxicity outcomes using a high-dose-rate (HDR) versus low-dose-rate (LDR) brachytherapy boost, Brachytherapy. 21 (2022) 599–604.
S. Nouranian, M. Ramezani, I. Spadinger, W.J. Morris, S.E. Salcudean, P. Abolmaesumi, Learning-based multi-label segmentation of transrectal ultrasound images for prostate brachytherapy, IEEE Trans. Med. Imaging. 35 (2016) 921–932.
X. Xu, T. Sanford, B. Turkbey, S. Xu, B.J. Wood, P. Yan, Shadow-consistent Semi-supervised learning for prostate ultrasound segmentation, IEEE Trans. Med. Imaging. 41 (2022) 1331–1345.
L. Rundo, C. Han, Y. Nagano, J. Zhang, R. Hataya, C. Militello, A. Tangherloni, M.S. Nobile, C. Ferretti, D. Besozzi, M.C. Gilardi, S. Vitabile, G. Mauri, H. Nakayama, P. Cazzaniga, USE-Net: Incorporating squeeze-and-excitation blocks into U-Net for prostate zonal segmentation of multi-institutional MRI datasets, Neurocomputing. 365 (2019) 31–43.
X. Yang, S. Zhan, D. Xie, H. Zhao, T. Kurihara, Hierarchical prostate MRI segmentation via level set clustering with shape prior, Neurocomputing. 257 (2017) 154–163.
A. Salimi, M.A. Pourmina, M.-S. Moin, Fully automatic prostate segmentation in MR images using a new hybrid active contour-based approach, Signal Image Video Process. 12 (2018) 1629–1637.
N. Orlando, I. Gyacskov, D.J. Gillies, F. Guo, C. Romagnoli, D. D’Souza, D.W. Cool, D.A. Hoover, A. Fenster, Effect of dataset size, image quality, and image type on deep learning-based automatic prostate segmentation in 3D ultrasound, Phys. Med. Biol. 67 (2022) 074002.
T. Peng, C. Tang, J. Wang, Prostate segmentation of ultrasound images based on interpretable-guided mathematical Model, in: Int. Conf. Multimed. Model. MMM, Springer, 2022: pp. 166–177.
T. Peng, C. Tang, Y. Wu, J. Cai, Semi-automatic prostate segmentation from ultrasound images using machine learning and principal curve based on interpretable mathematical model expression, Front. Oncol. 12 (2022).
S.M.S. Shah, S. Batool, I. Khan, M.U. Ashraf, S.H. Abbas, S.A. Hussain, Feature extraction through parallel Probabilistic Principal Component Analysis for heart disease diagnosis, Phys. Stat. Mech. Its Appl. 482 (2017) 796–807.
J. Zhang, W. Cui, X. Guo, B. Wang, Z. Wang, Classification of digital pathological images of non-Hodgkin’s lymphoma subtypes based on the fusion of transfer learning and principal component analysis, Med. Phys. 47 (2020) 4241–4253.
T. Hastie, W. Stuetzle, Principal Curves, J. Am. Stat. Assoc. 84 (1989) 502–516.
B. Kegl, T. Linder, K. Zeger, Learning and design of principal curves, IEEE Trans. Pattern Anal. Mach. Intell. 22 (2000) 281–297.
E.C. Correa Moraes, D.D. Ferreira, A principal curve-based method for data clustering, in: 2016 Int. Jt. Conf. Neural Netw. IJCNN, IEEE, Vancouver, BC, 2016: pp. 3966–3971.
J.J. Verbeek, N. Vlassis, B. Krose, A k-segments algorithm for finding principal curves, Pattern Recognit. Lett. 23 (2002) 1009–1017.
T. Peng, Y. Wang, T.C. Xu, L. Shi, J. Jiang, S. Zhu, Detection of lung contour with closed principal curve and machine learning, J. Digit. Imaging. 31 (2018) 520–533.
T. Peng, Y. Wang, T.C. Xu, X. Chen, Segmentation of lung in chest radiographs using hull and closed polygonal line method, IEEE Access. 7 (2019) 137794–137810.
T. Peng, C. Tang, Y. Wu, J. Cai, H-SegMed: A hybrid method for prostate segmentation in TRUS images via improved closed principal curve and improved enhanced machine learning, Int. J. Comput. Vis. 92 (2022).
G. Biau, A. Fischer, Parameter selection for principal curves, IEEE Trans. Inf. Theory. 58 (2012) 1924–1939.
Y. Guo, A. Şengür, Y. Akbulut, A. Shipley, An effective color image segmentation approach using neutrosophic adaptive mean shift clustering, Measurement. 119 (2018) 28–40.
M. R. Chen, B. P. Chen, G.-Q. Zeng, K. D. Lu, P. Chu, An adaptive fractional-order BP neural network based on extremal optimization for handwritten digits recognition, Neurocomputing. 391 (2020) 260–272.
E.C.C. Moraes, D.D. Ferreira, G.B. Vitor, B.H.G. Barbosa, Data clustering based on principal curves, Adv. Data Anal. Classif. 14 (2020) 77–96.
R. Wu, B. Wang, A. Xu, Functional data clustering using principal curve methods, Commun. Stat. - Theory Methods. (2021) 1–20.
S. Anand, S. Mittal, O. Tuzel, P. Meer, Semi-supervised kernel mean shift clustering, IEEE Trans. Pattern Anal. Mach. Intell. 36 (2014) 1201–1215.
B. Kégl, A. Krzyzak, Piecewise linear skeletonization using principal curves, IEEE Trans. Pattern Anal. Mach. Intell. 24 (2002) 59–74.
Y. Cheng, Mean shift, mode seeking, and clustering, IEEE Trans. Pattern Anal. Mach. Intell. 17 (1995) 790–799.
D. Comaniciu, V. Ramesh, P. Meer, The variable bandwidth mean shift and data-driven scale selection, in: Proc. Eighth IEEE Int. Conf. Comput. Vis. ICCV 2001, IEEE Comput. Soc, Vancouver, BC, Canada, 2001: pp. 438–445.
Y. Guo, A. Şengür, A novel image segmentation algorithm based on neutrosophic similarity clustering, Appl. Soft Comput. 25 (2014) 391–398.
N. Leema, H.K. Nehemiah, A. Kannan, Neural network classifier optimization using Differential Evolution with Global Information and Back Propagation algorithm for clinical datasets, Appl. Soft Comput. 49 (2016) 834–844.
M. Xiao, W.X. Zheng, G. Jiang, J. Cao, Undamped oscillations generated by hopf bifurcations in fractional-order recurrent neural networks with caputo derivative, IEEE Trans. Neural Netw. Learn. Syst. 26 (2015) 3201–3214.
L. Rice, E. Wong, J.Z. Kolter, Overfitting in adversarially robust deep learning, in: 2020: pp. 8093–8104.
B.L. Kalman, S.C. Kwasny, Why tanh: choosing a sigmoidal function, in: Proc. Int. Jt. Conf. Neural Netw., IEEE, Baltimore, MD, USA, 1992: pp. 578–581.
R. Hecht-Nielsen, Theory of the Backpropagation Neural Network, Neural Netw. Percept. (1992) 65–93.
N. Qian, On the momentum term in gradient descent learning algorithms, Neural Netw. 12 (1999) 145–151.
T. Peng, J. Zhao, Y. Gu, C. Wang, Y. Wu, X. Cheng, J. Cai, H-ProMed: ultrasound image segmentation based on the evolutionary neural network and an improved principal curve, Pattern Recognit. 131 (2022) 108890.
J. Wang, Y. Wen, Y. Gou, Z. Ye, H. Chen, Fractional-order gradient descent learning of BP neural networks with Caputo derivative, Neural Netw. 89 (2017) 19–30.
C. Bao, Y. Pu, Y. Zhang, Fractional-Order Deep Backpropagation Neural Network, Comput. Intell. Neurosci. 2018 (2018) 1–10.
T. Peng, Y. Gu, Z. Ye, X. Cheng, J. Wang, A-LugSeg: Automatic and explainability-guided multi-site lung detection in chest X-ray images, Expert Syst. Appl. 198 (2022) 116873.
T. Peng, T.C. Xu, Y. Wang, F. Li, Deep belief network and closed polygonal line for lung segmentation in chest radiographs, Comput. J. (2020).
N. Thapa, M. Chaudhari, S. McManus, K. Roy, R.H. Newman, H. Saigo, D.B. Kc, DeepSuccinylSite: a deep learning based approach for protein succinylation site prediction, BMC Bioinformatics. 21 (2020) 63.
T. Peng, C. Wang, Y. Zhang, J. Wang, H-SegNet: hybrid segmentation network for lung segmentation in chest radiographs using mask region-based convolutional neural network and adaptive closed polyline searching method, Phys. Med. Biol. 67 (2022) 075006.
D. Cashman, A. Perer, R. Chang, H. Strobelt, Ablate, Variate, and contemplate: visual analytics for discovering neural architectures, IEEE Trans. Vis. Comput. Graph. 26 (2019) 863–873.
Z. Zhou, M.M.R. Siddiquee, N. Tajbakhsh, J. Liang, UNet++: Redesigning skip connections to exploit multiscale features in image segmentation, IEEE Trans. Med. Imaging. 39 (2020) 1856–1867.
R. Zhao, B. Qian, X. Zhang, Y. Li, R. Wei, Y. Liu, Y. Pan, Rethinking Dice Loss for medical image segmentation, in: 2020 IEEE Int. Conf. Data Min. ICDM, IEEE, Sorrento, Italy, 2020: pp. 851–860.
Y. Lei, S. Tian, X. He, T. Wang, B. Wang, P. Patel, A.B. Jani, H. Mao, W.J. Curran, T. Liu, X. Yang, Ultrasound prostate segmentation based on multidirectional deeply supervised V-Net, Med. Phys. 46 (2019) 3194–3206.
Y. Wang, H. Dou, X. Hu, L. Zhu, X. Yang, M. Xu, J. Qin, P.-A. Heng, T. Wang, D. Ni, Deep attentive features for prostate segmentation in 3d transrectal ultrasound, IEEE Trans. Med. Imaging. 38 (2019) 2768–2778.
K.B. Girum, A. Lalande, R. Hussain, G. Créhange, A deep learning method for real-time intraoperative US image segmentation in prostate brachytherapy, Int. J. Comput. Assist. Radiol. Surg. 15 (2020) 1467–1476.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by Yiyun Wu, Jing Zhao, and Caishan Wang. The first draft of the manuscript was written by Tao Peng, and writing checking and review and supervision was performed by Jin Wang and Jing Cai. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics Approval
This is an observational study. The Research Ethics Committees of Jiangsu Province Hospital of Chinese Medicine, Beijing Tsinghua Changgung Hospital, and Second Affiliated Hospital of Soochow University have confirmed that no ethical approval is required.
Consent to Participate
Informed consent was obtained from all individual participants included in the study.
Consent for Publication
The authors affirm that human research participants provided informed consent for publication of all the images.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix
Appendix
-
Used symbols in this work
Appendix table used symbols in this work
Description | Symbols | |
|---|---|---|
Global variables | D-dimensional real number set space | RD |
Number of points in dataset | n | |
X-axis coordinate of data point | x | |
Y-axis coordinate of data point | y | |
ASPC | Principal curve | f |
Data point set/data point | Pn/pn | |
Vertex/segment subset of principal curve | Vi = {v1, v2, …, viv}/Si = {s1, s2, …, sis} | |
Vertex/segment of principal curve | v/s | |
Number of vertices/segments of principal curve | iv/is | |
Projection index | t | |
Mean shift vector | m(•) | |
Normalization factor | z | |
Kernel bandwidth | h | |
Radially symmetric kernel | K(·) | |
Kernel density estimator | S(·) | |
Derivative of the kernel profile | L(·) | |
Bright/indeterminate/non-bright pixel point sets | TC/IC /FC | |
Intensity value at data point p in the image | gd | |
Gradient value at data point p in the image | Gd | |
Kernel function of the indeterminacy filter | GIc | |
Standard deviation of kernel function | \(\sigma_{I}\) | |
Adjustment parameters in the linear function | ap1/ap2 | |
Average indeterminacy value of the current cluster point | Icavg | |
FBNNL | Number of neurons of input layer | I |
Number of neurons of hidden layer | H | |
Number of neurons of output layer | O | |
Weight from input layer to the hidden layer | w1 | |
Weight from hidden layer to the output layer | w2 | |
Threshold of the hidden neuron | a | |
Threshold of the output neuron | b | |
Learning rate from input layer to the hidden layer | μ1 | |
Learning rate from hidden layer to the output layer | μ2 | |
Adjustment parameter | ap | |
Fractional order | α | |
Iteration number | g | |
Output of output units | c(•) | |
Total error | E | |
Evaluation metrics | Dice similarity coefficient | DSC |
Jaccard similarity coefficient | OMG | |
Accuracy | ACC |
-
Values of important neural parameters
After training, we can determine the optimal parameters of our model to determine the experimental result contour according to Eqs. (14) and (15). Table 5 represents the values of important parameters of our model for expressing the contours in three randomly selected cases, and the qualitative results of these cases are shown in Fig. 8.
-
MSC method
To search the cluster of data, Cheng et al. [27] proposed the MSC method, whose details are summarized as follows:
Step 1: Compute the mean shift vector mh(pi), shown as follows.
To perform kernel density estimation, the kernel density estimator S(•) for pi is denoted as
where h is kernel bandwidth with a constant, K(·) is the radially symmetric kernel, and z is a normalization factor.
The density gradient estimator is obtained as the gradient of the density estimator based on Eq. (16), shown as below:
where L(p) = K' (p) shows the derivative of the selected kernel profile. The first term of Eq. (17) is a constant, and the second term of Eq. (17) is called the mean shift vector m(p), which points to the direction of the greatest increase in density, shown below:
Step 2: Update each initial point pi on the m(p) direction according to pi = + mh(pi).
Step 3: Iterate Step 1 and Step 2 until convergence.
Step 4: Determine the cluster point.
-
NAMSC method
According to the traditional MSC method, Guo et al. [21] proposed the NAMSC method, which combined an NS-based filter into the MSC method. The details of the NAMSC method are shown as follows:
Step 1: To unify the dataset, the initial data points Pn are normalized into the range of {(− 1, − 1) ~ (1,1)}.
Step 2: Map the Pn to each channel of the neutrosophic domain, where Tc(Pn), Ic(Pn), and Fc(Pn) represent the bright, indeterminate, and non-bright pixel point sets, respectively.
where gd(x, y) and Gd(x, y) are the intensity value and gradient value at the position of (x, y) in the image, respectively.
Step 3: Convolute each channel using the indeterminacy filter.
where \(\sigma_{I}\) is the standard deviation, which is used to determine the shape of the kernel function. GIc is a kernel function of the indeterminacy filter. ap1 and ap2 are the adjustment parameters in the linear function, which is used to transform the indeterminate value to filter’s parameter value.
Step 4: Compute the indeterminate values of the channels in the neutrosophic domain.
where Tc' is the result after indeterminate filter on Tc and fc is the filter size.
Step 5: Select a point pi (x, y) and compute the bandwidth h according to the corresponding indeterminate value.
where Icavg is the average indeterminacy value of the current cluster point. \(Tc^{\prime}_{\max}\) and \(Tc^{\prime}_{\min}\) are the maximum and minimum of Tc at the current cluster among all the channels of the neutrosophic domain, respectively.
Step 6: Compute the mean shift vector m(p) (same with Step 2 of the MSC method).
Step 7: Transfer each initial point pi on the m(p) direction (same with Step 3 of the MSC method).
Step 8: Go to Step 6 until it meets the condition \(\nabla S\left(p\right)=0\) (same as Step 4 of the MSC method).
Step 9: Update the bandwidth h using the average value of the indeterminate values in the current cluster.
Step 10: Go to Step 6 until the mean of the current cluster point become unchanged.
Step 11: Go to Step 5 until all the data points are clustered.
Step 12: Exit the loop and output the cluster points.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Peng, T., Wu, Y., Zhao, J. et al. Ultrasound Prostate Segmentation Using Adaptive Selection Principal Curve and Smooth Mathematical Model. J Digit Imaging 36, 947–963 (2023). https://doi.org/10.1007/s10278-023-00783-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-023-00783-3