Abstract
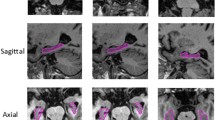
Precise segmentation of the hippocampus is essential for various human brain activity and neurological disorder studies. To overcome the small size of the hippocampus and the low contrast of MR images, a dual multilevel constrained attention GAN for MRI-based hippocampus segmentation is proposed in this paper, which is used to provide a relatively effective balance between suppressing noise interference and enhancing feature learning. First, we design the dual-GAN backbone to effectively compensate for the spatial information damage caused by multiple pooling operations in the feature generation stage. Specifically, dual-GAN performs joint adversarial learning on the multiscale feature maps at the end of the generator, which yields an average Dice coefficient (DSC) gain of 5.95% over the baseline. Next, to suppress MRI high-frequency noise interference, a multilayer information constraint unit is introduced before feature decoding, which improves the sensitivity of the decoder to forecast features by 5.39% and effectively alleviates the network overfitting problem. Then, to refine the boundary segmentation effects, we construct a multiscale feature attention restraint mechanism, which forces the network to concentrate more on effective multiscale details, thus improving the robustness. Furthermore, the dual discriminators D1 and D2 also effectively prevent the negative migration phenomenon. The proposed DMCA-GAN obtained a DSC of 90.53% on the Medical Segmentation Decathlon (MSD) dataset with tenfold cross-validation, which is superior to the backbone by 3.78%.

















Similar content being viewed by others
Data Availability
The dataset is public and can be downloaded from http://medicaldecathlon.com/.
Code Availability
The code used in this work is available from the first author upon request.
References
Anand KS, Dhikav V (2012) Hippocampus in health and disease: An overview. Annals of Indian Academy of Neurology 15(4):239
Abuhmed T, El-Sappagh S, Alonso JM (2021) Robust hybrid deep learning models for alzheimer’s progression detection. Knowledge-Based Systems 213:106,688
Du A, Schuff N, Amend D, et al (2001) Magnetic resonance imaging of the entorhinal cortex and hippocampus in mild cognitive impairment and alzheimer’s disease. Journal of Neurology, Neurosurgery & Psychiatry 71(4):441–447
Cendes F, Andermann F, Gloor P, et al (1993) Mri volumetric measurement of amygdala and hippocampus in temporal lobe epilepsy. Neurology 43(4):719–719
Perez SM, Shah A, Asher A, et al (2013) Hippocampal deep brain stimulation reverses physiological and behavioural deficits in a rodent model of schizophrenia. International Journal of Neuropsychopharmacology 16(6):1331–1339
Sahay A, Hen R (2007) Adult hippocampal neurogenesis in depression. Nature neuroscience 10(9):1110–1115
Sheynin S, Wolf L, Ben-Zion Z, et al (2021) Deep learning model of fmri connectivity predicts ptsd symptom trajectories in recent trauma survivors. Neuroimage 238:118,242
Broadbent NJ, Squire LR, Clark RE (2004) Spatial memory, recognition memory, and the hippocampus. Proceedings of the National Academy of Sciences 101(40):14,515–14,520
Schenck JF (1996) The role of magnetic susceptibility in magnetic resonance imaging: Mri magnetic compatibility of the first and second kinds. Medical physics 23(6):815–850
Antonelli M, Reinke A, Bakas S, et al (2022) The medical segmentation decathlon. Nature communications 13(1):4128
Simpson AL, Antonelli M, Bakas S, et al (2019) A large annotated medical image dataset for the development and evaluation of segmentation algorithms. arXiv preprint arXiv:190209063
Iglesias JE, Sabuncu MR (2015) Multi-atlas segmentation of biomedical images: a survey. Medical image analysis 24(1):205–219
Minaee S, Boykov Y, Porikli F, et al (2021) Image segmentation using deep learning: A survey. IEEE transactions on pattern analysis and machine intelligence 44(7):3523–3542
Thyreau B, Sato K, Fukuda H, et al (2018) Segmentation of the hippocampus by transferring algorithmic knowledge for large cohort processing. Medical image analysis 43:214–228
Guo Y, Wu Z, Shen D (2020) Learning longitudinal classification-regression model for infant hippocampus segmentation. Neurocomputing 391:191–198
Wu Z, Gao Y, Shi F, et al (2018) Segmenting hippocampal subfields from 3t mri with multi-modality images. Medical image analysis 43:10–22
Van Essen DC, Smith SM, Barch DM, et al (2013) The wu-minn human connectome project: an overview. Neuroimage 80:62–79
Ataloglou D, Dimou A, Zarpalas D, et al (2019) Fast and precise hippocampus segmentation through deep convolutional neural network ensembles and transfer learning. Neuroinformatics 17(4):563–582
Brusini I, Lindberg O, Muehlboeck JS, et al (2020) Shape information improves the cross-cohort performance of deep learning-based segmentation of the hippocampus. Frontiers in neuroscience 14:15
Khalvati F, Salmanpour A, Rahnamayan S, et al (2016) Sequential registration-based segmentation of the prostate gland in mr image volumes. Journal of digital imaging 29(2):254–263
Nobakht S, Schaeffer M, Forkert ND, et al (2021) Combined atlas and convolutional neural network-based segmentation of the hippocampus from mri according to the adni harmonized protocol. Sensors 21(7):2427
Roy AG, Conjeti S, Navab N, et al (2019) Quicknat: A fully convolutional network for quick and accurate segmentation of neuroanatomy. NeuroImage 186:713–727
Son J, Park SJ, Jung KH (2019) Towards accurate segmentation of retinal vessels and the optic disc in fundoscopic images with generative adversarial networks. Journal of digital imaging 32(3):499–512
Yang Z, Zhuang X, Mishra V, et al (2020) Cast: A multi-scale convolutional neural network based automated hippocampal subfield segmentation toolbox. NeuroImage 218:116,947
Zandifar A, Fonov V, Coupé P, et al (2017) A comparison of accurate automatic hippocampal segmentation methods. NeuroImage 155:383–393
Madhumalini M, Devi TM (2022) Detection of glaucoma from fundus images using novel evolutionary-based deep neural network. Journal of Digital Imaging pp 1–15
Barzegar Z, Jamzad M (2022) An efficient optimization approach for glioma tumor segmentation in brain mri. Journal of Digital Imaging pp 1–14
Mecheter I, Alic L, Abbod M, et al (2020) Mr image-based attenuation correction of brain pet imaging: review of literature on machine learning approaches for segmentation. Journal of Digital Imaging 33(5):1224–1241
Shahedi M, Cool DW, Bauman GS, et al (2017) Accuracy validation of an automated method for prostate segmentation in magnetic resonance imaging. Journal of digital imaging 30(6):782–795
Öztürk Ş, Özkaya U (2020) Skin lesion segmentation with improved convolutional neural network. Journal of digital imaging 33(4):958–970
Ngo TA, Lu Z, Carneiro G (2017) Combining deep learning and level set for the automated segmentation of the left ventricle of the heart from cardiac cine magnetic resonance. Medical image analysis 35:159–171
Liu M, Li F, Yan H, et al (2020) A multi-model deep convolutional neural network for automatic hippocampus segmentation and classification in alzheimer’s disease. Neuroimage 208:116,459
Jack Jr CR, Bernstein MA, Fox NC, et al (2008) The alzheimer’s disease neuroimaging initiative (adni): Mri methods. Journal of Magnetic Resonance Imaging: An Official Journal of the International Society for Magnetic Resonance in Medicine 27(4):685–691
Hazarika RA, Maji AK, Syiem R, et al (2022) Hippocampus segmentation using u-net convolutional network from brain magnetic resonance imaging (mri). Journal of Digital Imaging pp 1–17
Ranem A, González C, Mukhopadhyay A (2022) Continual hippocampus segmentation with transformers. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp 3711–3720
Dosovitskiy A, Beyer L, Kolesnikov A, et al (2020) An image is worth 16x16 words: Transformers for image recognition at scale. arXiv preprint arXiv:201011929
Isensee F, Jaeger PF, Kohl SA, et al (2021) nnu-net: a self-configuring method for deep learning-based biomedical image segmentation. Nature methods 18(2):203–211
Lin M, Cai Q, Zhou J (2022) 3d md-unet: A novel model of multi-dataset collaboration for medical image segmentation. Neurocomputing 492:530–544
Chen H, Qin Z, Ding Y, et al (2020) Brain tumor segmentation with deep convolutional symmetric neural network. Neurocomputing 392:305–313
Harms J, Lei Y, Wang T, et al (2019) Paired cycle-gan-based image correction for quantitative cone-beam computed tomography. Medical physics 46(9):3998–4009
Isola P, Zhu JY, Zhou T, et al (2017) Image-to-image translation with conditional adversarial networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 1125–1134
Li M, Tang H, Chan MD, et al (2020) Dc-al gan: pseudoprogression and true tumor progression of glioblastoma multiform image classification based on dcgan and alexnet. Medical Physics 47(3):1139–1150
Zhang X, Yang Y, Li T, et al (2021) Cmc: a consensus multi-view clustering model for predicting alzheimer’s disease progression. Computer Methods and Programs in Biomedicine 199:105,895
Shi Y, Cheng K, Liu Z (2019) Hippocampal subfields segmentation in brain mr images using generative adversarial networks. Biomedical engineering online 18(1):1–12
Chen Y, Yang X, Cheng K, et al (2020) Efficient 3d neural networks with support vector machine for hippocampus segmentation. In: 2020 international conference on artificial intelligence and computer engineering (ICAICE), IEEE, pp 337–341
Han C, Rundo L, Murao K, et al (2019) Gan-based multiple adjacent brain mri slice reconstruction for unsupervised alzheimer’s disease diagnosis. In: International Meeting on Computational Intelligence Methods for Bioinformatics and Biostatistics, Springer, pp 44–54
Yu W, Lei B, Ng MK, et al (2021) Tensorizing gan with high-order pooling for alzheimer’s disease assessment. IEEE Transactions on Neural Networks and Learning Systems
Li M, Lian F, Guo S (2022) Multi-scale selection and multi-channel fusion model for pancreas segmentation using adversarial deep convolutional nets. Journal of Digital Imaging 35(1):47–55
Luo Y, Liu P, Guan T, et al (2019) Significance-aware information bottleneck for domain adaptive semantic segmentation. In: Proceedings of the IEEE/CVF International Conference on Computer Vision, pp 6778–6787
Luo Y, Zheng L, Guan T, et al (2019) Taking a closer look at domain shift: Category-level adversaries for semantics consistent domain adaptation. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp 2507–2516
Jain A, Nandakumar K, Ross A (2005) Score normalization in multimodal biometric systems. Pattern recognition 38(12):2270–2285
Otsu N (1979) A threshold selection method from gray-level histograms. IEEE transactions on systems, man, and cybernetics 9(1):62–66
Song S, Zheng Y, He Y (2017) A review of methods for bias correction in medical images. Biomedical Engineering Review 1(1)
Plassard AJ, Bao S, McHugo M, et al (2021) Automated, open-source segmentation of the hippocampus and amygdala with the open vanderbilt archive of the temporal lobe. Magnetic resonance imaging 81:17–23
First MB (2005) Structured clinical interview for dsm-iv-tr axis i disorders. (No Title)
Pruessner JC, Li LM, Serles W, et al (2000) Volumetry of hippocampus and amygdala with high-resolution mri and three-dimensional analysis software: minimizing the discrepancies between laboratories. Cerebral cortex 10(4):433–442
Woolard AA, Heckers S (2012) Anatomical and functional correlates of human hippocampal volume asymmetry. Psychiatry Research: Neuroimaging 201(1):48–53
Yushkevich PA, Piven J, Cody Hazlett H, et al (2006) User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. Neuroimage 31(3):1116–1128
Zhang G, Yang Z, Huo B, et al (2021) Automatic segmentation of organs at risk and tumors in ct images of lung cancer from partially labelled datasets with a semi-supervised conditional nnu-net. Computer methods and programs in biomedicine 211:106,419
Cao L, Li L, Zheng J, et al (2018) Multi-task neural networks for joint hippocampus segmentation and clinical score regression. Multimedia Tools and Applications 77(22):29,669–29,686
Porter E, Fuentes P, Siddiqui Z, et al (2020) Hippocampus segmentation on noncontrast ct using deep learning. Medical physics 47(7):2950–2961
Xie L, Wisse LE, Wang J, et al (2023) Deep label fusion: A generalizable hybrid multi-atlas and deep convolutional neural network for medical image segmentation. Medical Image Analysis 83:102,683
He Y, Yang D, Roth H, et al (2021) Dints: Differentiable neural network topology search for 3d medical image segmentation. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp 5841–5850
Funding
This work was supported in part by the National Natural Science Foundation of China (Grant No. 61976126) and Shandong Nature Science Foundation of China (Grant No. ZR2019 MF003).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation and data collection and analysis were performed by Xue Chen, Yanjun Peng, Dapeng Li and Jindong Sun. The first draft of the manuscript was written by Xue Chen, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Consent to Participate
Informed consent was obtained from all individual participants included in the study. The external dataset 2018 Medical Segmentation Decathlon challenge is available in the MSD repository; all data are downloadable from http://medicaldecathlon.com/.
Consent for Publication
All data were made available online under Creative Commons license CC-BY-SA 4.0, allowing the data to be shared or redistributed in any format and improved upon, with no commercial restrictions. Under this license, the appropriate credit must be given (by citation to this paper [50]), with a link to the license and any changes noted. The images can be redistributed under the same license.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, X., Peng, Y., Li, D. et al. DMCA-GAN: Dual Multilevel Constrained Attention GAN for MRI-Based Hippocampus Segmentation. J Digit Imaging 36, 2532–2553 (2023). https://doi.org/10.1007/s10278-023-00854-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-023-00854-5