Abstract
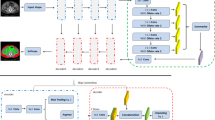
Automatic segmentation of adipose tissue from CT images is an essential module of medical assistant diagnosis. A large scale of abdominal cross-section CT images can be used to segment subcutaneous adipose tissue (SAT) and visceral adipose tissue (VAT) with deep learning method. However, the CT images still need to be professionally and accurately annotated to improve the segmentation quality. The paper proposes a semi-supervised segmentation network based on adversarial learning. The model is called URO-GAN and consists of two paths used to segment SAT and VAT, respectively. An SAT-to-VAT transmission mechanism is set up between these two paths, where several inverse-SAT excitation blocks are set to help the SAT segmentation network guide the VAT segmentation network. An untrustworthy region optimization mechanism is proposed to improve the segmentation quality and keep the adversarial learning stable. With the confidence map output from the discriminator network, an optimizer network is used to fix the error in the masks predicted by the segmentation network. The URO-GAN achieves good results by training with 84 annotated images and 3969 unannotated images. Experimental results demonstrate the effectiveness of our approach on the segmentation of adipose tissue in medical images.













Similar content being viewed by others
References
GBD 2015 Obesity Collaborators (2017) Health effects of overweight and obesity in 195 countries over 25 years. N Engl J Med 377(1):13–27
Wu Y, Jiang X, Fang Z, Gao Y, Fujita H (2021) Multi-modal 3D object detection by 2D-guided precision anchor proposal and multi-layer fusion. Appl Soft Comput 108:107405
Zhu K, Jiang X, Fang Z, Gao Y, Fujita H, Hwang JN (2021) Photometric transfer for direct visual odometry. Knowl-Based Syst 213:106671
Mourtzakis M, Prado CM, Lieffers JR, Reiman T, McCargar LJ, Baracos VE (2008) A practical and precise approach to quantification of body composition in cancer patients using computed tomography images acquired during routine care. Appl Physiol Nutrit Metabolism 33(5):997–1006
Després JP, Lemieux I (2006) Abdominal obesity and metabolic syndrome. Nature 444 (7121):881–887
De Larochellière E, Côté J, Gilbert G, Bibeau K, Ross MK, Dion-Roy V, ..., Larose É (2014) Visceral/epicardial adiposity in nonobese and apparently healthy young adults: association with the cardiometabolic profile. Atherosclerosis 234(1):23–29
Long J, Shelhamer E, Darrell T (2015) Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 3431–3440
Ronneberger O, Fischer P, Brox T (2015) U-net: Convolutional networks for biomedical image segmentation. In: International Conference on Medical image computing and computer-assisted intervention. Springer, Cham, pp 234–241
Jégou S, Drozdzal M, Vazquez D, Romero A, Bengio Y (2017) The one hundred layers tiramisu: Fully convolutional densenets for semantic segmentation. In: Proceedings of the IEEE conference on computer vision and pattern recognition workshops, pp 11–19
Cui H, Yuwen C, Jiang L, Xia Y, Zhang Y (2021) Multiscale attention guided U-Net architecture for cardiac segmentation in short-axis MRI images. Comput Methods Prog Biomed 106142
Fu J, Liu J, Tian H, Li Y, Bao Y, Fang Z, Lu H (2019) Dual attention network for scene segmentation. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp 3146–3154
Hu J, Shen L, Sun G (2018) Squeeze-and-excitation networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 7132–7141
Roy AG, Navab N, Wachinger C (2018) Concurrent spatial and channel ’squeeze & excitation’ in fully convolutional networks. In: International conference on medical image computing and computer-assisted intervention. Springer, Cham, pp 421–429
Sinha A, Dolz J (2020) Multi-scale self-guided attention for medical image segmentation. IEEE J Biomed Health Inform
Li C, Tan Y, Chen W, Luo X, He Y, Gao Y, Li F (2020) ANU-Net: Attention-based Nested U-Net to exploit full resolution features for medical image segmentation. Comput Graph 90:11–20
Sun J, Darbehani F, Zaidi M, Wang B (2020) SAUNet: shape attentive U-Net for interpretable medical image segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Cham, pp 797–806
Estrada S, Lu R, Conjeti S, Orozco-Ruiz X, Panos-Willuhn J, Breteler MM, Reuter M (2020) Fatsegnet: A fully automated deep learning pipeline for adipose tissue segmentation on abdominal dixon MRI. Magnet Resonance Med 83(4):1471–1483
Dabiri S, Popuri K, Ma C, Chow V, Feliciano EMC, Caan BJ, ..., Beg MF (2020) Deep learning method for localization and segmentation of abdominal CT. Comput Med Imaging Graph 85:101776
Pan S, Hou X, Li H, Sheng B, Fang R, Xue Y, ..., Qin J (2019) Abdominal Adipose Tissue Segmentation in MRI with Double Loss Function Collaborative Learning. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Cham, pp 41–49
Sadananthan SA, Prakash B, Leow MKS, Khoo CM, Chou H, Venkataraman K, ..., Velan SS (2015) Automated segmentation of visceral and subcutaneous (deep and superficial) adipose tissues in normal and overweight men. J Magnet Reson Imag 41(4):924–934
Choi YJ, Seo YK, Lee EJ, Chung YS (2015) Quantification of visceral fat using dual-energy x-ray absorptiometry and its reliability according to the amount of visceral fat in Korean adults. J Clin Densitom 18(2):192–197
Feng Z, Nie D, Wang L, Shen D (2018) Semi-supervised learning for pelvic MR image segmentation based on multi-task residual fully convolutional networks. In: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018). IEEE, pp 885–888
Bai W, Oktay O, Sinclair M, Suzuki H, Rajchl M, Tarroni G, ..., Rueckert D (2017) Semi-supervised learning for network-based cardiac MR image segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Cham, pp 253–260
Nie D, Gao Y, Wang L, Shen D (2018) ASDNet: attention based semi-supervised deep networks for medical image segmentation. In: International conference on medical image computing and computer-assisted intervention. Springer, Cham, pp 370–378
Goodfellow IJ, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, ..., Bengio Y (2014) Generative adversarial networks. arXiv:1406.2661
Luc P, Couprie C, Chintala S, Verbeek J (2016) Semantic Segmentation using Adversarial Networks. In: NIPS Workshop on Adversarial Training
Zhang Y, Yang L, Chen J, Fredericksen M, Hughes DP, Chen DZ (2017) Deep adversarial networks for biomedical image segmentation utilizing unannotated images. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Cham, pp 408–416
Han L, Huang Y, Dou H, Wang S, Ahamad S, Luo H, ..., Zhang J (2020) Semi-supervised segmentation of lesion from breast ultrasound images with attentional generative adversarial network. Comput Methods Programs Biomed 189:105275
Hung WC, Tsai YH, Liou YT, Lin YY, Yang MH (2019) Adversarial learning for semi-supervised semantic segmentation. In: 29th British Machine Vision Conference BMVC , p 2018
Li G, Wan J, He S, Liu Q, Ma B (2020) Semi-supervised semantic segmentation using adversarial learning for pavement crack detection. IEEE Access 8:51446–51459
Decourt C, Duong L (2020) Semi-supervised generative adversarial networks for the segmentation of the left ventricle in pediatric MRI. Comput Biol Med 123:103884
Roy AG, Siddiqui S, Pölsterl S, Navab N, Wachinger C (2020) Squeeze, excite guided few-shot segmentation of volumetric images. Med Image Anal 59:101587
Shaban A, Bansal S, Liu Z, Essa I, Boots B (2017) One-shot learning for semantic segmentation. arXiv:1709.03410
Fei-Fei L, Fergus R, Perona P (2006) One-shot learning of object categories. IEEE Trans Pattern Anal Machine Intell 28(4):594–611
Miller EG, Matsakis NE, Viola PA (2000) Learning from one example through shared densities on transforms. In: Proceedings IEEE Conference on Computer Vision and Pattern Recognition. CVPR 2000 (Cat. No. PR00662), vol 1. IEEE, pp 464– 471
Fei-Fei L (2006) Knowledge transfer in learning to recognize visual objects classes. In: Proceedings of the International Conference on Development and Learning (ICDL), p 11
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 4700–4708
Lee CY, Xie S, Gallagher P, Zhang Z, Tu Z (2015) Deeply-supervised nets. In: Artificial intelligence and statistics. PMLR, pp 562–570
Mao X, Li Q, Xie H, Lau RY, Wang Z, Paul Smolley S (2017) Least squares generative adversarial networks. In: Proceedings of the IEEE international conference on computer vision, pp 2794–2802
Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, ..., Zheng X (2016) Tensorflow: A system for large-scale machine learning. In: 12th USENIX symposium on operating systems design and implementation (OSDI), vol 16, pp 265–283
Kingma DP, Ba J (2014) Adam: A method for stochastic optimization. arXiv:1412.6980
Dice LR (1945) Measures of the amount of ecologic association between species. Ecol 26(3):297–302
Holger R, Lu L, Seff A, Cherry KM, Hoffman J, Wang Sn, ..., Summers RM (2015) A new 2.5 D representation for lymph node detection in CT. The Cancer Imaging Archive. https://doi.org/10.7937/K9/TCIA.2015.AQIIDCNM
Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, ..., Prior F (2013) The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imag 26 (6):1045–1057
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interests
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Shen, K., Quan, H., Han, J. et al. URO-GAN: An untrustworthy region optimization approach for adipose tissue segmentation based on adversarial learning. Appl Intell 52, 10247–10269 (2022). https://doi.org/10.1007/s10489-021-02976-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10489-021-02976-1