Abstract
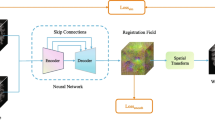
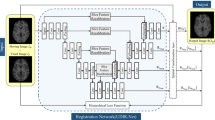
Medical image registration is a vital initial step in medical image analysis that aims to establish accurate correspondence between image pairs. Various methods have achieved impressive results on this task over the last few years. However, the existing methods fail to achieve desirable registration accuracy because they do not pay sufficient attention on the integration between global and local information in medical images and ignore the geometric constraints in anatomical regions. The registration results still need to be refined to become sufficiently accurate for clinical application considering the current situation. In this paper, we propose a novel part-and-whole registration framework for 3D medical images. The proposed framework consists of two encoder-decoder architectures which take image pairs and patch pairs as inputs, respectively, and estimate the deformation field as the result. In particular, we propose a multi-view feature slicing attention module and embed it into the part-and-whole architecture to enhance the context relation between the local and global information. A voxel-shape-aware loss function is developed to refine the registration performance. We validate the proposed framework on several 3D human brain magnetic resonance image datasets. Results show that our method consistently outperforms recent state-of-the-art baselines.









Similar content being viewed by others
References
Ahmad S, Khan M (2015) Deformable image registration based on elastodynamics. Mach Vis Appl 26:689–710
Ashburner J (2007) A fast diffeomorphic image registration algorithm. Neuroimage 38(1):95–113
Avants BB, Epstein CL, Grossman M, Gee JC (2008) Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal 12(1):26–41
Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC (2011) A reproducible evaluation of ants similarity metric performance in brain image registration. Neuroimage 54(3):2033–2044
Balakrishnan G, Zhao A, Sabuncu MR, Guttag J, Dalca AV (2019) Voxelmorph: a learning framework for deformable medical image registration. IEEE Trans Med Imaging 38(8):1788–1800
Beg MF, Miller MI, Trouvé A, Younes L (2005) Computing large deformation metric mappings via geodesic flows of diffeomorphisms. Int J Comput Vis 61(2):139–157
Broit C (1981) Optimal registration of deformed images. University of Pennsylvania, Pennsylvania
Cao X, Yang J, Zhang J, Nie D, Kim M, Wang Q, Shen D (2017) Deformable image registration based on similarity-steered cnn regression. In: International conference on medical image computing and computer-assisted intervention. Springer, pp 300–308
Cao X, Yang J, Zhang J, Wang Q, Yap PT, Shen D (2018) Deformable image registration using a cue-aware deep regression network. IEEE Trans Biomed Eng 65(9):1900–1911
Chen J, He Y, Frey EC, Li Y, Du Y (2021) Vit-v-net: Vision transformer for unsupervised volumetric medical image registration. arXiv:210406468
Chen X, Williams BM, Vallabhaneni SR, Czanner G, Williams R, Zheng Y (2019) Learning active contour models for medical image segmentation. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 11632–11640
Coevoet E, Reynaert N, Lartigau E, Schiappacasse L, Dequidt J, Duriez C (2015) Registration by interactive inverse simulation: application for adaptive radiotherapy. Int J Comput Assist Radiol Surg 10(8):1193–1200
Di Martino A, Yan CG, Li Q, Denio E, Castellanos FX, Alaerts K, Anderson JS, Assaf M, Bookheimer SY, Dapretto M et al (2014) The autism brain imaging data exchange: towards a large-scale evaluation of the intrinsic brain architecture in autism. Mol Psychiatry 19(6):659–667
Dice LR (1945) Measures of the amount of ecologic association between species. Ecol 26(3):297–302
Fan J, Cao X, Yap PT, Shen D (2019) Birnet: Brain image registration using dual-supervised fully convolutional networks. Med Image Anal 54:193–206
Fischl B (2012) Freesurfer. Neuroimage 62(2):774–781
Fu Y, Lei Y, Wang T, Curran WJ, Liu T, Yang X (2020) Deep learning in medical image registration: a review. Physics in Medicine & Biology 65(20):20TR01
Hoopes A, Hoffmann M, Fischl B, Guttag J, Dalca AV (2021) Hypermorph: Amortized hyperparameter learning for image registration. In: International conference on information processing in medical imaging. Springer, pp 3–17
Hou Q, Zhou D, Feng J (2021) Coordinate attention for efficient mobile network design. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 13713–13722
Hu J, Shen L, Sun G (2018) Squeeze-and-excitation networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 7132–7141
Huang W, Yang H, Liu X, Li C, Zhang I, Wang R, Zheng H, Wang S (2021) A coarse-to-fine deformable transformation framework for unsupervised multi-contrast mr image registration with dual consistency constraint. IEEE Transactions on Medical Imaging
Jaderberg M, Simonyan K, Zisserman A, et al. (2015) Spatial transformer networks. Adv Neural Inf Process Syst 28:2017–2025
Jingya Z, Jiajun W, Xiuying W, Dagan F (2013) The adaptive fem elastic model for medical image registration. Phys Med Biol 59(1):97
Kingma DP, Ba J (2014) Adam: A method for stochastic optimization. arXiv:14126980
Liu Z, Huang J, Zhu C, Peng X, Du X (2021) Residual attention network using multi-channel dense connections for image super-resolution. Appl Intell 51(1):85–99
Lu E, Hu X (2021) Image super-resolution via channel attention and spatial attention. Appl Intell 1–9
Ma Y, Niu D, Zhang J, Zhao X, Yang B, Zhang C (2021) Unsupervised deformable image registration network for 3d medical images. Appl Intell 1–14
Marcus DS, Wang TH, Parker J, Csernansky JG, Morris JC, Buckner RL (2007) Open access series of imaging studies (oasis): cross-sectional mri data in young, middle aged, nondemented, and demented older adults. J Cogn Neurosci 19(9):1498–1507
Miao S, Wang ZJ, Zheng Y, Liao R (2016) Real-time 2d/3d registration via cnn regression. In: 2016 IEEE 13Th international symposium on biomedical imaging (ISBI). IEEE, pp 1430–1434
Milham MP, Fair D, Mennes M, Mostofsky SH et al (2012) The adhd-200 consortium: a model to advance the translational potential of neuroimaging in clinical neuroscience. Front Syst Neurosci 6:62
Mok TC, Chung A (2020) Fast symmetric diffeomorphic image registration with convolutional neural networks. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 4644–4653
Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack CR, Jagust W, Trojanowski JQ, Toga AW, Beckett L (2005) Ways toward an early diagnosis in alzheimer’s disease: the alzheimer’s disease neuroimaging initiative (adni). Alzheimer’s & Dementia 1(1):55–66
Paszke A, Gross S, Chintala S, Chanan G, Yang E, DeVito Z, Lin Z, Desmaison A, Antiga L, Lerer A (2017) Automatic differentiation in pytorch
Ronneberger O, Fischer P, Brox T (2015) U-net: Convolutional networks for biomedical image segmentation. In: International Conference on Medical image computing and computer-assisted intervention. Springer, pp 234–241
Rueckert D, Sonoda LI, Hayes C, Hill DL, Leach MO, Hawkes DJ (1999) Nonrigid registration using free-form deformations: application to breast mr images. IEEE Trans Med Imaging 18(8):712–721
Shen Z, Vialard FX, Niethammer M (2019) Region-specific diffeomorphic metric mapping. Adv Neural Inf Process Syst 32
Siebert H, Hansen L, Heinrich MP (2021) Fast 3d registration with accurate optimisation and little learning for learn2reg 2021. arXiv:211203053
Sokooti H, De Vos B, Berendsen F, Lelieveldt BP, Išgum I, Staring M (2017) Nonrigid image registration using multi-scale 3d convolutional neural networks. In: International conference on medical image computing and computer-assisted intervention. Springer, pp 232–239
de Vos BD, Berendsen FF, Viergever MA, Staring M, Išgum I (2017) End-to-end unsupervised deformable image registration with a convolutional neural network. In: Deep learning in medical image analysis and multimodal learning for clinical decision support. Springer, pp 204–212
de Vos BD, Berendsen FF, Viergever MA, Sokooti H, Staring M, Išgum I (2019) A deep learning framework for unsupervised affine and deformable image registration. Med Image Anal 52:128–143
Wang C, Deng C, Ivanov V (2020) Sag-vae: End-To-end joint inference of data representations and feature relations. In: 2020 International joint conference on neural networks (IJCNN). IEEE, pp 1–9
Woo S, Park J, Lee JY, Kweon IS (2018) Cbam: Convolutional block attention module. In: Proceedings of the European conference on computer vision (ECCV), pp 3–19
Zhang W, Zhao Y (2021) Hierarchical registration of brain images based on b-splines and laplacian commutators. Optik 241:167022
Zhang Z, Sejdić E (2019) Radiological images and machine learning: trends, perspectives, and prospects. Comput Biol Med 108:354–370
Zhao S, Dong Y, Chang EI, Xu Y et al (2019) Recursive cascaded networks for unsupervised medical image registration. In: Proceedings of the IEEE/CVF international conference on computer vision, pp 10600–10610
Acknowledgements
This research was supported by Natural Science Foundation of Shandong province (Nos. ZR2019MF013, ZR2019BF 026), Project of Jinan Scientific Research Leaderś Laboratory (No. 2018GXRC023).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendices
Appendix A: Detailed of the neural network
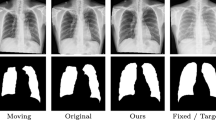
Appendix B: Comparison of different methods on vital anatomical regions
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, J., Liu, Z., Ma, Y. et al. Part-and-whole: A novel framework for deformable medical image registration. Appl Intell 53, 16630–16647 (2023). https://doi.org/10.1007/s10489-022-04329-y
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10489-022-04329-y