Abstract
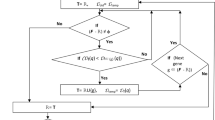
This study explores gene selection in a single cell gene decision space (scgd-space) based on class-consistent technology and fuzzy rough iterative computation model (FRIC-model). Gene expression data (ge-data) exhibit characteristics such as limited sample size, high dimensionality, and noise. Due to their high dimensionality, gene selection must be carried out before clustering and classifying them. The existing gene selection methods based on equivalence relation are not effective for ge-data owing to the strictness of the equality between gene expression values. In order to overcome this weakness, class-consistent technology of replacing equality with approximate equality between gene expression values is first proposed. Then, “the class consistency between gene expression values is fed back to the gene set” is considered with the help of class-consistent technology, and fuzzy symmetric relations on the cell set of a scgd-space are induced. In addition, fuzzy rough approximations in a scgd-space are defined. Next, FRIC-model is given. This model employs the iterative computation strategy to define fuzzy rough approximations and dependency functions. A gene selection algorithm based on this model is designed. Finally, the designed algorithm is testified in several publicly open ge-data sets to estimate its performance. The experimental results show that the designed algorithm is more effective than some existing algorithms.










Similar content being viewed by others
References
Benesty J, Chen J, Huang Y, Cohen I (2009) Pearson correlation coefficient, Noise reduction in speech processing, Springer, pp. 1–4
Biase F, Cao X, Zhong S (2014) Cell fate inclination within 2-cell and 4-cell mouse embryos revealed by single-cell RNA sequencing. Genome Res 24:1787–1796
Buettner F, Natarajan KN, Casale FP, Proserpio V, Scialdone A, Theis FJ, Teichmann SA, Marioni JC, Stegle O (2015) Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat Biotechnol 33:1–8
Bommert A, Welchowski T, Schmid M, Rahnenf\(\ddot{u}\)hrer J (2022) Benchmark of filter methods for feature selection in high-dimensional gene expression survival data. Brief Bioinform 23:bbab354
Cornelis C, Jensen R, Martin GH, Slezak D (2010) Attribute selection with fuzzy decision reducts. Inf Sci 180:209–224
Demisar J (2006) Statistical comparisons of classifiers over multiple data sets. J Mach Learn Res 7:1–30
Derrac J, Garc\(\acute{i}\)a S, Molina D, Herrera F, (2011) A practical tutorial on the use of nonparametric statistical tests as a methodology for comparing evolutionary and swarm intelligence algorithms. Swarm Evol Comput 1:3–18
Dai JH, Hu H, Wu WZ, Qian YH, Huang DB (2018) Maximal-discernibility-pair-based approach to attribute reduction in fuzzy rough sets. IEEE Trans Fuzzy Syst 26(4):2175–2187
Deng Q, Ramskld D, Reinius B, Sandberg R (2014) Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science 343:193–196
Engel I, Seumois G, Chavez L, Samaniego-Castruita D, White B, Chawla A, Mock D, Vijayanand P, Kronenberg M (2016) Innate-like functions of natural killer T cell subsets result from highly divergent gene programs. Nat Immunol 17:728C739
Friedman M (1940) A comparison of alternative tests of significance for the problem of m rankings. Annals Math Stat 11:86–92
Fan X, Zhang X, Wu X, Guo H, Hu Y, Tang F, Huang Y (2015) Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome Biol 16(148):1–17
Gao L, Cai MJ, Li QG (2023) A relative granular ratio-based outlier detection method in heterogeneous data. Inf Sci 622:710–731
Guyon I, Elisseeff A (2003) An introduction to variable and feature selection. J Mach Learn Res 3:1157–1182
Grover A, Sanjuan-Pla A, Thongjuea S, Carrelha J, Giustacchini A, Gambardella A, Macaulay I, Mancini E, Luis TC, Mead A (2016) Single-cell RNA sequencing reveals molecular and functional platelet bias of aged haematopoietic stem cells. Nat Commun 7:11075–11075
Huang D, Chen YY, Liu F, Li ZW (2023) Feature selection for multiset-valued data based on fuzzy conditional information entropy using iterative model and matrix operation. Appl Soft Comput 142:110345
Kolodziejczyk AA, Kim JK, Tsang JC, Ilicic T, Henriksson J, Natarajan KN, Tuck AC, Gao X, Bıhler M, Liu P (2015) Single cell RNA-sequencing of pluripotent states unlocks modular transcriptional variation. Cell Stem Cell 17:471–485
Kimmerling RJ, Szeto GL, Li JW, Genshaft AS, Kazer SW, Payer KR, de Riba Borrajo J, Blainey PC, Irvine DJ, Shalek AK (2016) A microfluidic platform enabling single-cell RNA-seq of multigenerational lineages. Nat Commun 7:1–7
Leng N, Chu L, Barry C, Li Y, Choi J, Li X, Jiang P, Stewart RM, Thomson JA, Kendziorski C (2015) Oscope identifies oscillatory genes in unsynchronized single-cell RNA-seq experiments. Nat Methods 12:947C950
Li X, Cui X, Wang J, Wang Y, Li Y, Wang L, Wan H, Li T, Feng G, Shuai L (2016) Generation and application of mouse-rat allodiploid embryonic stem cells. Cell 164:279–292
Li Z, Feng J, Zhang J, Liu F, Wang P, Wen C (2022) Gaussian kernel based gene selection in a single cell gene decision space. Inf Sci 610:1029–1057
Li ZW, Liu XF, Dai JH, Chen JL, Fujita H (2020) Measures of uncertainty based on Gaussian kernel for a fully fuzzy information system. Knowl-Based Syst 196:105791
Li ZW, Qu LD, Zhang GQ, Xie NX (2021) Attribute selection for heterogeneous data based on information entropy. Int J Gen Syst 50(5):548–566
Li ZW, Zhang PF, Ge X, Xie NX, Zhang GQ, Wen CF (2019) Uncertainty measurement for a fuzzy relation information system. IEEE Trans Fuzzy Syst 27(12):2338–2352
Meng ZQ, Shi ZZ (2009) A fast approach to attribute reduction in incomplete decision systems with tolerance relation-based rough sets. Inf Sci 179:2774–2793
Mwangi B, Tian TS, Soares JC (2014) A review of feature reduction techniques in neuroimaging. Neuroinformatics 12:229–244
Pawlak Z (1982) Rough sets. Int J Comput Inform Sci 11:341–356
Pawlak Z (1991) Rough sets: theoretical aspects of reasoning about data. Kluwer Academic Publishers, Dordrecht
Robnik-\(\check{S}\)ikonja M, Kononenko I, (2003) Theoretical and empirical analysis of ReliefF and RReliefF. Mach Learn 53:23–69
\(\breve{S}\)id\(\acute{a}\)k Z, (1967) Rectangular confidence regions for the means of multivariate normal distributions. J Am Stat Assoc 62:626–633
Sheng J, Li WV (2021) Selecting gene features for unsupervised analysis of single-cell gene expression data. Brief Bioinform 22:bbab295
Sharma A, Rani R (2019) C-HMOSHSSA: gene selection for cancer classification using multi-objective meta-heuristic and machine learning methods. Comput Methods Prog Biomed 178:219–235
Singh S, Shreevastava S, Som T, Somani G (2020) A fuzzy similarity-based rough set approach for attribute selection in set-valued information systems. Soft Comput 24(6):4675–4691
Sun L, Zhang XY, Qian YH, Xu JC, Zhang SG, Tian Y (2019) Joint neighborhood entropy-based gene selection method with fisher score for tumor classification. Appl Intell 49:1245–1259
Treutlein B, Brownfield DG, Wu AR, Neff NF, Mantalas GL, Espinoza FH, Desai TJ, Krasnow MA, Quake SR (2014) Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature 509:371–375
Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL (2014) The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 32:381–386
Trabelsi S, Elouedi Z (2010) Heuristic method for attribute selection from partially uncertain data using rough sets. Int J Gen Syst 39(3):271–290
Tabakhi S, Moradi P, Akhlaghian F (2014) An unsupervised feature selection algorithm based on ant colony optimization. Eng Appl Artif Intell 32:112–123
Ting DT, Wittner BS, Ligorio M, Jordan NV, Shah AM, Miyamoto DT, Aceto N, Bersani F, Brannigan BW, Xega K (2014) Single-cell RNA sequencing identifies extracellular matrix gene expression by pancreatic circulating tumor cells. Cell Rep 8:1905–1918
Wang YB, Chen XJ, Dong K (2019) Attribute reduction via local conditional entropy. Int J Mach Learn Cybern 10(12):3619–3634
Wang CZ, Huang Y, Shao MW, Hu QH, Chen DG (2020) Feature selection based on neighborhood self-information. IEEE Trans Cybern 50:4031–4042
Wang CZ, Wang Y, Shao MW, Qian YH, Chen DG (2020) Fuzzy rough attribute reduction for categorical data. IEEE Trans Fuzzy Syst 28(5):818–830
Xu F, Cai MJ, Song H, Dai JH (2022) The selection of feasible strategies based on consistency measurement of cliques. Inf Sci 583:33–55
Yang D, Cai MJ, Li QG, Xu F (2022) Multigranulation fuzzy probabilistic rough set model on two universes. Int J Approx Reason 145:18–35
Yang Y, Huh R, Houston WC, Lin Y, Michael IL, Li Y (2019) SAFE-clustering: single-cell aggregated (from ensemble) clustering for single-cell RNA-seq data. Bioinforma 35:1269–1277
Yang W, Wang K, Zuo W (2012) Neighborhood component feature selection for high-dimensional data. J Comput 7:161–168
Yao YY, Zhang XY (2017) Class-specific attribute reducts in rough set theory. Inf Sci 418–419:601–618
Zadeh LA (1965) Fuzzy sets. Inf. Control 8:338–356
Zhang J, Zhang GQ, Li ZW, Qu LD, Wen CF (2021) Feature selection in a neighborhood decision information system with application to single cell RNA data classification. Appl Soft Comput 113:107876
Acknowledgements
The authors would like to thank the editors and the anonymous reviewers for their valuable comments and suggestions, which have helped immensely in improving the quality of the paper. This work is supported by Guangxi First-class Discipline Statistics Construction Project Fund, Natural Science Foundation of Guangxi Province (2021GXNSFAA220076, 2021GXNSFAA220114), Key Fields Project of Universities in Guangdong Province (2023ZDZX1063, 2023ZDZX1065, 2021ZDZX4109) and Scientific Research Platform of Guangdong Songshan Polytechnic (2022xjkypt02).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflicts of interest
All authors declare that there is no conflict of interests regarding the publication of this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, J., Yu, G., Huang, D. et al. Gene selection in a single cell gene decision space based on class-consistent technology and fuzzy rough iterative computation model. Appl Intell 53, 30113–30132 (2023). https://doi.org/10.1007/s10489-023-05115-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10489-023-05115-0