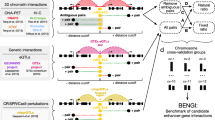
Abstract
In mammalian and vertebrate genomes, the promoter regions of the gene and their distal enhancers may be located millions of base-pairs from each other, while a promoter may not interact with the closest enhancer. Since base-pair proximity is not a good indicator of these interactions, there is a significant body of work to develop methods for understanding Enhancer-Promoter Interactions (EPI) from genetic and epigenomic marks. Over the last decade, several machine learning and deep learning methods have reported increasingly higher accuracies for predicting EPI. Typically, these approaches perform analysis by randomly splitting the dataset of Enhancer-Promoter (EP) pairs into training and testing subsets followed by model training. However, the aforementioned random splitting inadvertently causes information leakage by assigning EP pairs from the same genomic region to both testing and training sets. As a result, it has been pointed out in the literature that the performance of EPI prediction algorithms is overestimated because of genomic region overlap among the training and testing parts of the data. Building on that, in this paper we propose to use a more thorough training and testing paradigm i.e., Leave-one-chromosome-out (LOCO) cross-validation for EPI prediction. LOCO has been used in other bioinformatics contexts and ensures that there is no genomic overlap between training and testing sets enabling more fair estimation of performance. We demonstrate that a deep learning algorithm which gives higher accuracies when trained and tested on random-splitting setting, drops drastically in performance under LOCO setting, showing overestimation of performance in previous literature. We also propose a novel hybrid multi-branch neural network architecture for EPI prediction. In particular, our architecture has one branch consisting of a deep neural network, while the other branch extracts traditional k-mer features derived from the nucleotide sequence. The two branches are later merged and the neural network is trained jointly to force the network to learn feature representations which are already not covered by k-mer features. We show that the hybrid architecture performs significantly better in a realistic and fair LOCO testing paradigm, demonstrating it can learn more general aspects of EP interactions instead of overfitting to genomic regions. Through this paper we are also releasing the LOCO splitting-based EPI dataset to encourage other research groups to benchmark their EPI algorithms using a consistent LOCO paradigm. Research data is available in this public repository: https://github.com/malikmtahir/EPI





Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Data availability
Research data and code are available in this public repository: https://github.com/malikmtahir/EPI
References
Mora A, Sandve GK, Gabrielsen OS, Eskeland R (2016) In the loop: promoter-enhancer interactions and bioinformatics. Brief Bioinform 17(6):980–995
Talukder A, Saadat S, Li X, Hu H (2019) Epip: a novel approach for condition-specific enhancer-promoter interaction prediction. Bioinformatics 35(20):3877–3883
Cai X, Hou L, Su N, Hu H, Deng M, Li X (2010) Systematic identification of conserved motif modules in the human genome. BMC Genomics 11:1–10
Zhang Y, Wong C-H, Birnbaum RY, Li G, Favaro R, Ngan CY, Lim J, Tai E, Poh HM, Wong E (2013) Chromatin connectivity maps reveal dynamic promoter-enhancer long-range associations. Nature 504(7479):306–310
Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin DU, Jung I, Wu H, Zhai Y, Tang Y (2015) Crispr inversion of ctcf sites alters genome topology and enhancer/promoter function. Cell 162(4):900–910
Singh S, Yang Y, Póczos B, Ma J (2019) Predicting enhancer-promoter interaction from genomic sequence with deep neural networks. Quantitative Biology 7:122–137
Panigrahi A, O’Malley BW (2021) Mechanisms of enhancer action: the known and the unknown. Genome Biol 22:1–30
Huang C, Helin K (2023) Catching active enhancers via h2b n-terminal acetylation. Nature Genetics 1–2
Lettice LA, Heaney SJ, Purdie LA, Li L, Beer P, Oostra BA, Goode D, Elgar G, Hill RE, Graaff E (2003) A long-range shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet 12(14):1725–1735
Mills C, Marconett CN, Lewinger JP, Mi H (2023) Peacock: a machine learning approach to assess the validity of cell type-specific enhancer-gene regulatory relationships. npj Systems Biology and Applications 9(1):9
Panigrahi AK, Lonard DM, O’Malley BW (2023) Enhancer-promoter entanglement explains their transcriptional interdependence. Proc Natl Acad Sci 120(4):2216436120
Williamson I, Hill RE, Bickmore WA (2011) Enhancers: from developmental genetics to the genetics of common human disease. Dev Cell 21(1):17–19
Achinger-Kawecka J, Clark SJ (2017) Disruption of the 3d cancer genome blueprint. Epigenomics 9(1):47–55
Smemo S, Campos LC, Moskowitz IP, Krieger JE, Pereira AC, Nobrega MA (2012) Regulatory variation in a tbx5 enhancer leads to isolated congenital heart disease. Hum Mol Genet 21(14):3255–3263
Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES (2014) A 3d map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159(7):1665–1680
Javierre BM, Burren OS, Wilder SP, Kreuzhuber R, Hill SM, Sewitz S, Cairns J, Wingett SW, Várnai C, Thiecke MJ (2016) Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell 167(5):1369–1384
Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J (2012) Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 148(1):84–98
Belokopytova PS, Nuriddinov MA, Mozheiko EA, Fishman D, Fishman V (2020) Quantitative prediction of enhancer-promoter interactions. Genome Res 30(1):72–84
Whalen S, Truty RM, Pollard KS (2016) Enhancer-promoter interactions are encoded by complex genomic signatures on looping chromatin. Nat Genet 48(5):488–496
Buckle A, Brackley CA, Boyle S, Marenduzzo D, Gilbert N (2018) Polymer simulations of heteromorphic chromatin predict the 3d folding of complex genomic loci. Mol Cell 72(4):786–797
Chiariello AM, Annunziatella C, Bianco S, Esposito A, Nicodemi M (2016) Polymer physics of chromosome large-scale 3d organisation. Sci Rep 6(1):29775
Di Pierro M, Cheng RR, Lieberman Aiden E, Wolynes PG, Onuchic JN (2017) De novo prediction of human chromosome structures: Epigenetic marking patterns encode genome architecture. Proc Natl Acad Sci 114(46):12126–12131
Chen Y, Wang Y, Xuan Z, Chen M, Zhang MQ (2016) De novo deciphering three-dimensional chromatin interaction and topological domains by wavelet transformation of epigenetic profiles. Nucleic Acids Res 44(11):106–106
Zeng W, Wu M, Jiang R (2018) Prediction of enhancer-promoter interactions via natural language processing. BMC Genomics 19:13–22
Mao W, Kostka D, Chikina M (2017) Modeling enhancer-promoter interactions with attention-based neural networks. bioRxiv, 219667
Zhuang Z, Shen X, Pan W (2019) A simple convolutional neural network for prediction of enhancer-promoter interactions with dna sequence data. Bioinformatics 35(17):2899–2906
Hong Z, Zeng X, Wei L, Liu X (2020) Identifying enhancer-promoter interactions with neural network based on pre-trained dna vectors and attention mechanism. Bioinformatics 36(4):1037–1043
Jing F, Zhang S-W, Zhang S (2020) Prediction of enhancer-promoter interactions using the cross-cell type information and domain adversarial neural network. BMC Bioinformatics 21(1):1–16
Liu S, Xu X, Yang Z, Zhao X, Liu S, Zhang W (2021) Epihc: Improving enhancer-promoter interaction prediction by using hybrid features and communicative learning. IEEE/ACM Trans Comput Biol Bioinf 19(6):3435–3443
Fan Y, Peng B (2022) Stackepi: identification of cell line-specific enhancer-promoter interactions based on stacking ensemble learning. BMC Bioinformatics 23(1):272
Min X, Ye C, Liu X, Zeng X (2021) Predicting enhancer-promoter interactions by deep learning and matching heuristic. Brief Bioinform 22(4):254
Ahmed FS, Aly S, Liu X (2024) Epi-trans: an effective transformer-based deep learning model for enhancer promoter interaction prediction. BMC Bioinformatics 25(1):216
Su W, Xie X-Q, Liu X-W, Gao D, Ma C-Y, Zulfiqar H, Yang H, Lin H, Yu X-L, Li Y-W (2023) irna-ac4c: a novel computational method for effectively detecting n4-acetylcytidine sites in human mrna. Int J Biol Macromol 227:1174–1181
Guo S-H, Deng E-Z, Xu L-Q, Ding H, Lin H, Chen W, Chou K-C (2014) inuc-pseknc: a sequence-based predictor for predicting nucleosome positioning in genomes with pseudo k-tuple nucleotide composition. Bioinformatics 30(11):1522–1529
Chen W, Feng P-M, Lin H, Chou K-C (2013) irspot-psednc: identify recombination spots with pseudo dinucleotide composition. Nucleic Acids Res 41(6):68–68
Lin H, Deng E-Z, Ding H, Chen W, Chou K-C (2014) ipro54-pseknc: a sequence-based predictor for identifying sigma-54 promoters in prokaryote with pseudo k-tuple nucleotide composition. Nucleic Acids Res 42(21):12961–12972
Kabir M, Hayat M (2016) irspot-gaensc: identifing recombination spots via ensemble classifier and extending the concept of chou’s pseaac to formulate dna samples. Mol Genet Genomics 291:285–296
Tahir M, Hayat M (2016) inuc-stnc: a sequence-based predictor for identification of nucleosome positioning in genomes by extending the concept of saac and chou’s pseaac. Mol BioSyst 12(8):2587–2593
Feng C-Q, Zhang Z-Y, Zhu X-J, Lin Y, Chen W, Tang H, Lin H (2019) iterm-pseknc: a sequence-based tool for predicting bacterial transcriptional terminators. Bioinformatics 35(9):1469–1477
DeLong ER, DeLong DM, Clarke-Pearson DL (1988) Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 837–845
Khosla P, Teterwak P, Wang C, Sarna A, Tian Y, Isola P, Maschinot A, Liu C, Krishnan D (2020) Supervised contrastive learning. Adv Neural Inf Process Syst 33:18661–18673
Gunel B, Du J, Conneau A, Stoyanov V (2020) Supervised contrastive learning for pre-trained language model fine-tuning. arXiv:2011.01403
Liu X, Song C, Huang F, Fu H, Xiao W, Zhang W (2022) Graphcdr: a graph neural network method with contrastive learning for cancer drug response prediction. Brief Bioinform 23(1):457
Lin S, Chen W, Chen G, Zhou S, Wei D-Q, Xiong Y (2022) Mddi-scl: predicting multi-type drug-drug interactions via supervised contrastive learning. Journal of Cheminformatics 14(1):1–12
Heinzinger M, Littmann M, Sillitoe I, Bordin N, Orengo C, Rost B (2022) Contrastive learning on protein embeddings enlightens midnight zone. NAR genomics and bioinformatics 4(2):043
Rajadhyaksha N, Chitkara A (2023) Graph contrastive learning for multi-omics data. arXiv:2301.02242
Lee H, Ozbulak U, Park H, Depuydt S, De Neve W, Vankerschaver J (2024) Assessing the reliability of point mutation as data augmentation for deep learning with genomic data. BMC Bioinformatics 25(1):170
Chen J, Mowlaei ME, Shi X (2020) Population-scale genomic data augmentation based on conditional generative adversarial networks. In: Proceedings of the 11th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics, pp. 1–6
Dinsdale NK, Jenkinson M, Namburete AI (2021) Deep learning-based unlearning of dataset bias for mri harmonisation and confound removal. Neuroimage 228:117689
Ashraf A, Khan S, Bhagwat N, Chakravarty M, Taati B (2018) Learning to unlearn: Building immunity to dataset bias in medical imaging studies. Machine Learning for Health Workshop, NeurIPS, Canada
Khan SS, Shen Z, Sun H, Patel A, Abedi A (2022) Supervised contrastive learning for detecting anomalous driving behaviours from multimodal videos. In: 2022 19th Conference on Robots and Vision (CRV), pp. 16–23. IEEE
Lin JC-W, Shao Y, Djenouri Y, Yun U (2021) Asrnn: A recurrent neural network with an attention model for sequence labeling. Knowl-Based Syst 212:106548
Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser Ł, Polosukhin I (2017) Attention is all you need. Advances in neural information processing systems 30
Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, Bengio Y (2020) Generative adversarial networks. Commun ACM 63(11):139–144
Radford A, Narasimhan K, Salimans T, Sutskever I et al (2018) Improving language understanding by generative pre-training
Strokach A, Kim PM (2022) Deep generative modeling for protein design. Curr Opin Struct Biol 72:226–236
Luo R, Sun L, Xia Y, Qin T, Zhang S, Poon H, Liu T-Y (2022) Biogpt: generative pre-trained transformer for biomedical text generation and mining. Brief Bioinform 23(6):409
Byrd JB, Greene AC, Prasad DV, Jiang X, Greene CS (2020) Responsible, practical genomic data sharing that accelerates research. Nat Rev Genet 21(10):615–629
Schwab AP, Luu HS, Wang J, Park JY (2018) Genomic privacy. Clin Chem 64(12):1696–1703
Health U (2015) Genomic Data Sharing: A Two-Part Series. https://osp.od.nih.gov/genomic-data-sharing-a-two-part-series
Acknowledgements
Financial support from the following funding agencies is acknowledged: • Canadian Institutes of Health Research(CIHR) • Japan Agency for Medical Research and Development (AMED)
Author information
Authors and Affiliations
Contributions
A.A. and M.T. conceived the idea, designed experiments, and data analysis. S.K. contributed to implementation of the experiments and simulations. J.D. and S.Y contributed to revised and edited the manuscript and provided suggestions. All authors analyzed the results and made critical changes on the manuscript at all stages.
Corresponding author
Ethics declarations
Conflict of interest/Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tahir, M., Khan, S.S., Davie, J. et al. LOCO-EPI: Leave-one-chromosome-out (LOCO) as a benchmarking paradigm for deep learning based prediction of enhancer-promoter interactions. Appl Intell 55, 71 (2025). https://doi.org/10.1007/s10489-024-05848-6
Accepted:
Published:
DOI: https://doi.org/10.1007/s10489-024-05848-6