Abstract
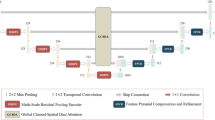
Medical image segmentation in clinical applications is important and challenging. Learning contextual features including local and global information is pivotal in effective medical image segmentation. Existing methods based on convolutional neural networks (CNNs) are usually constrained by limited receptive fields, resulting in inaccurate segmentation when dealing with local similarities and large-scale variations in complex medical images. In this paper, we focus on combining convolution and attention mechanisms for effective and efficient medical image segmentation, and propose a local and global contextual encoding network (LGCE-Net), which contains an encoder, a feature extraction module and a decoder. In the encoder, multi-scale feature maps are obtained through convolution and pooling operations. For feature extraction, the Dense Atrous Convolution Attention (DACA) block is introduced, which leverages atrous convolutions with different atrous rates and space-related attention to capture local information. Additionally, the Spatial Grid Attention (SGA) block, which combines grid attention and spatial attention to extract global contextual information, is presented to enhance the feature representations. Finally, sub-pixel convolution is used to restore semantic features extracted from the encoder and the feature extraction module. We conducted experiments on three public datasets and our approach outperforms CNN-based, attention-based and state-of-the-art CNN-Attention combined models. Moreover, our model runs at 200 Frames-Per-Second (FPS) when only 9.22M parameters are used. Our code will be released once the manuscript is accepted for publication.




Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Data Availability
The dataset used during the current study is publicly available, and the available links have been given in the manuscript.
References
Huo X, Sun G, Tian S, Wang Y, Yu L, Long J, Zhang W, Li A (2024) Hifuse: Hierarchical multi-scale feature fusion network for medical image classification. Biomed Signal Process Control 87:105534
Karaman A, Karaboga D, Pacal I, Akay B, Basturk A, Nalbantoglu U, Coskun S, Sahin O (2023) Hyper-parameter optimization of deep learning architectures using artificial bee colony (abc) algorithm for high performance real-time automatic colorectal cancer (crc) polyp detection. Appl Intell 53(12):15603–15620
Karaman A, Pacal I, Basturk A, Akay B, Nalbantoglu U, Coskun S, Sahin O, Karaboga D (2023) Robust real-time polyp detection system design based on yolo algorithms by optimizing activation functions and hyper-parameters with artificial bee colony (abc). Expert Syst Appl 221:119741
Wang R, Lei T, Cui R, Zhang B, Meng H, Nandi AK (2022) Medical image segmentation using deep learning: A survey. IET Image Proc 16(5):1243–1267
Kirillov A, Mintun E, Ravi N, Mao H, Rolland C, Gustafson L, Xiao T, Whitehead S, Berg AC, Lo W-Y et al (2023) Segment anything. arXiv:2304.02643
Zhang Y, Shen Z, Jiao R (2024) Segment anything model for medical image segmentation: Current applications and future directions. Comput Biol Med pp 108238
Ji G-P, Fan D-P, Xu P, Cheng M-M, Zhou B, Van Gool L (2023) Sam struggles in concealed scenes–empirical study on" segment anything". arXiv:2304.06022
Ji W, Li J, Bi Q, Li W, Cheng L (2023) Segment anything is not always perfect: An investigation of sam on different real-world applications. arXiv:2304.05750
Huang Y, Yang X, Liu L, Zhou H, Chang A, Zhou X, Chen R, Yu J, Chen J, Chen C et al (2023) Segment anything model for medical images? Med Image Anal pp 103061
Chen L-C, Papandreou G, Kokkinos I, Murphy K, Yuille AL (2017) Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans Pattern Anal Mach Intell 40(4):834–848
Shen C, Nguyen D, Zhou Z, Jiang SB, Dong B, Jia X (2020) An introduction to deep learning in medical physics: advantages, potential, and challenges. Phys Med Biol 65(5):05–01
Long J, Shelhamer E, Darrell T (2015) Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 3431–3440
Ronneberger O, Fischer P, Brox T (2015) U-net: Convolutional networks for biomedical image segmentation. In: Medical image computing and computer-assisted intervention–MICCAI 2015: 18th International Conference, Munich, Germany, October 5–9, 2015, Proceedings, Part III 18, Springer, pp 234–241
Yao W, Bai J, Liao W, Chen Y, Liu M, Xie Y (2024) From cnn to transformer: A review of medical image segmentation models. J Imaging Inform Med pp 1–19
Lin A, Chen B, Xu J, Zhang Z, Lu G, Zhang D (2022) Ds-transunet: Dual swin transformer u-net for medical image segmentation. IEEE Trans Instrum Meas 71:1–15
Liu Z, Mao H, Wu C.-Y, Feichtenhofer C, Darrell T, Xie S (2022) A convnet for the 2020s. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition pp 11976–11986
Gu Z, Cheng J, Fu H, Zhou K, Hao H, Zhao Y, Zhang T, Gao S, Liu J (2019) Ce-net: Context encoder network for 2d medical image segmentation. IEEE Trans Med Imaging 38(10):2281–2292
Guan S, Khan AA, Sikdar S, Chitnis PV (2019) Fully dense unet for 2-d sparse photoacoustic tomography artifact removal. IEEE J Biomed Health Inform 24(2):568–576
Xiao X, Lian S, Luo Z, Li S (2018) Weighted res-unet for high-quality retina vessel segmentation. In: 2018 9th International conference on information technology in medicine and education (ITME), IEEE, pp 327–331
Zhou Z, Rahman Siddiquee MM, Tajbakhsh N, Liang J (2018) Unet++: A nested u-net architecture for medical image segmentation. In: Deep learning in medical image analysis and multimodal learning for clinical decision support: 4th international workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, September 20, 2018, Proceedings 4, pp 3–11. Springer
Isensee F, Petersen J, Klein A, Zimmerer D, Jaeger P.F, Kohl S, Wasserthal J, Koehler G, Norajitra T, Wirkert S et al (2018) nnu-net: Self-adapting framework for u-net-based medical image segmentation. arXiv:1809.10486
Xie S, Girshick R, Dollár P, Tu Z, He K (2017) Aggregated residual transformations for deep neural networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 1492–1500
Bai C, Sun F, Zhang J, Song Y, Chen S (2022) Rainformer: Features extraction balanced network for radar-based precipitation nowcasting. IEEE Geosci Remote Sens Lett 19:1–5
Chan S, Yu M, Chen Z, Mao J, Bai C (2023) Regional contextual information modeling for small object detection on highways. IEEE Transactions on Instrumentation and Measurement
Brauwers G, Frasincar F (2021) A general survey on attention mechanisms in deep learning. IEEE Trans Knowl Data Eng
Li Z, Zheng Y, Shan D, Yang S, Li Q, Wang B, Zhang Y, Hong Q, Shen D (2024) Scribformer: Transformer makes cnn work better for scribble-based medical image segmentation. IEEE Trans Med Imaging
Chaoyang Z, Shibao S, Wenmao H, Pengcheng Z (2024) Fdr-transunet: A novel encoder-decoder architecture with vision transformer for improved medical image segmentation. Comput Biol Med 169:107858
Hu J, Shen L, Sun G (2018) Squeeze-and-excitation networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 7132–7141
Woo S, Park J, Lee J-Y, Kweon IS (2018) Cbam: Convolutional block attention module. In: Proceedings of the European conference on computer vision (ECCV), pp 3–19
Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez A.N, Kaiser Ł, Polosukhin I (2017) Attention is all you need. Adv Neural Inform Process Syst 30
Li J, Liu K, Hu Y, Zhang H, Heidari AA, Chen H, Zhang W, Algarni AD, Elmannai H (2023) Eres-unet++: Liver ct image segmentation based on high-efficiency channel attention and res-unet++. Comput Biol Med 158:106501
Chen B, Liu Y, Zhang Z, Lu G, Kong AWK (2023) Transattunet: Multi-level attention-guided u-net with transformer for medical image segmentation. IEEE Transactions on Emerging Topics in Computational Intelligence
Dosovitskiy A, Beyer L, Kolesnikov A, Weissenborn D, Zhai X, Unterthiner T, Dehghani M, Minderer M, Heigold G, Gelly S et al (2020) An image is worth 16x16 words: Transformers for image recognition at scale. arXiv:2010.11929
Chen J, Lu Y, Yu Q, Luo X, Adeli E, Wang Y, Lu L, Yuille AL, Zhou Y (2021) Transunet: Transformers make strong encoders for medical image segmentation. arXiv:2102.04306
Cao H, Wang Y, Chen J, Jiang D, Zhang X, Tian Q, Wang M (2023) Swin-unet: Unet-like pure transformer for medical image segmentation. In: Computer Vision–ECCV 2022 Workshops: Tel Aviv, Israel, October 23–27, 2022, Proceedings, Part III, Springer, pp 205–218
Lin X, Yan Z, Yu L, Cheng K-T (2022) C2ftrans: Coarse-to-fine transformers for medical image segmentation. arXiv:2206.14409
Gao Y, Zhou M, Metaxas DN (2021) Utnet: a hybrid transformer architecture for medical image segmentation. In: Medical image computing and computer assisted intervention–MICCAI 2021: 24th international conference, Strasbourg, France, September 27–October 1, 2021, Proceedings, Part III 24, Springer, pp 61–71
Yuan L, Hou Q, Jiang Z, Feng J, Yan S (2022) Volo: Vision outlooker for visual recognition. IEEE Trans Pattern Anal Mach Intell
Tu Z, Talebi H, Zhang H, Yang F, Milanfar P, Bovik A, Li Y (2022) Maxvit: Multi-axis vision transformer. In: Computer Vision–ECCV 2022: 17th European Conference, Tel Aviv, Israel, October 23–27, 2022, Proceedings, Part XXIV, Springer, pp 459–479
Yu F, Koltun V (2015) Multi-scale context aggregation by dilated convolutions. arXiv:1511.07122
Touvron H, Cord M, Douze M, Massa F, Sablayrolles A, Jégou H (2021) Training data-efficient image transformers & distillation through attention. In: International Conference on Machine Learning, PMLR, pp 10347–10357
Liu Z, Lin Y, Cao Y, Hu H, Wei Y, Zhang Z, Lin S, Guo B (2021) Swin transformer: Hierarchical vision transformer using shifted windows. In: Proceedings of the IEEE/CVF international conference on computer vision, pp 10012–10022
Guo J, Han K, Wu H, Tang Y, Chen X, Wang Y, Xu C (2022) Cmt: Convolutional neural networks meet vision transformers. In: Proceedings of the IEEE/CVF Conference on computer vision and pattern recognition, pp 12175–12185
Wang X, Shao M, Guo D, Cui Y, Huang X, Xia M, Bai C (2023) Multi-stage aggregation transformer for medical image segmentation. In: ICASSP 2023-2023 IEEE international conference on acoustics, speech and signal processing (ICASSP), IEEE, pp 1–5
Deng Y, Meng Y, Chen J, Yue A, Liu D, Chen J (2023) Tchange: A hybrid transformer-cnn change detection network. Remote Sensing 15(5):1219
Zhou H.-Y, Guo J, Zhang Y, Yu L, Wang L, Yu Y (2021) nnformer: Interleaved transformer for volumetric segmentation. aarXiv:2109.03201
Zhu J, Sheng Y, Cui H, Ma J, Wang J, Xi H (2023) Cross pyramid transformer makes u-net stronger in medical image segmentation. Biomed Signal Process Control 86:105361
Heidari M, Kazerouni A, Soltany M, Azad R, Aghdam E.K, Cohen-Adad J, Merhof D (2023) Hiformer: Hierarchical multi-scale representations using transformers for medical image segmentation. In: Proceedings of the IEEE/CVF winter conference on applications of computer vision, pp 6202–6212
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 4700–4708
Stergiou A, Poppe R (2022) Adapool: Exponential adaptive pooling for information-retaining downsampling. IEEE Trans Image Process 32:251–266
Kaiser L, Gomez AN, Chollet F (2017) Depthwise separable convolutions for neural machine translation. arXiv:1706.03059
Shi W, Caballero J, Huszár F, Totz J, Aitken AP, Bishop R, Rueckert D, Wang Z (2016) Real-time single image and video super-resolution using an efficient sub-pixel convolutional neural network. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 1874–1883
Staal J, Abràmoff MD, Niemeijer M, Viergever MA, Van Ginneken B (2004) Ridge-based vessel segmentation in color images of the retina. IEEE Trans Med Imaging 23(4):501–509
Kumar N, Verma R, Anand D, Zhou Y, Onder OF, Tsougenis E, Chen H, Heng P-A, Li J, Hu Z et al (2019) A multi-organ nucleus segmentation challenge. IEEE Trans Med Imaging 39(5):1380–1391
Kumar N, Verma R, Sharma S, Bhargava S, Vahadane A, Sethi A (2017) A dataset and a technique for generalized nuclear segmentation for computational pathology. IEEE Trans Med Imaging 36(7):1550–1560
Wang X, Yuan Y, Guo D, Huang X, Cui Y, Xia M, Wang Z, Bai C, Chen S (2022) Ssa-net: Spatial self-attention network for covid-19 pneumonia infection segmentation with semi-supervised few-shot learning. Med Image Anal 79:102459
Valanarasu JMJ, Patel VM (2022) Unext: Mlp-based rapid medical image segmentation network. In: Medical image computing and computer assisted intervention–MICCAI 2022: 25th International Conference, Singapore, September 18–22, 2022, Proceedings, Part V, Springer, pp 23–33
Chen Y, Dai X, Chen D, Liu M, Dong X, Yuan L, Liu Z (2022) Mobile-former: Bridging mobilenet and transformer. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 5270–5279
Tragakis A, Kaul C, Murray-Smith R, Husmeier D (2023) The fully convolutional transformer for medical image segmentation. In: Proceedings of the IEEE/CVF winter conference on applications of computer vision, pp 3660–3669
Xu Q, Ma Z, Na H, Duan W (2023) Dcsau-net: A deeper and more compact split-attention u-net for medical image segmentation. Comput Biol Med 154:106626
Chen J, Kao S-h, He H, Zhuo W, Wen S, Lee C-H, Chan S-HG (2023) Run, don’t walk: Chasing higher flops for faster neural networks. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 12021–12031
Lucchi A, Smith K, Achanta R, Knott G, Fua P (2011) Supervoxel-based segmentation of mitochondria in em image stacks with learned shape features. IEEE Trans Med Imaging 31(2):474–486
Wang X, Shao M, Guo D, Cui Y, Huang X, Xia M, Bai C (2023) Multi-stage aggregation transformer for medical image segmentation. In: ICASSP 2023-2023 IEEE international conference on acoustics, speech and signal processing (ICASSP), IEEE, pp 1–5
Acknowledgements
This work was supported in part by the National Natural Science Foundation of China under grant 62273308, in part by the Natural Science Foundation of Zhejiang Province under grants LY23F030007 and LY20H180006, in part by the Zhejiang Provincial Research Project on the Application of Public Welfare Technologies under grant LGF22F020023.
Author information
Authors and Affiliations
Contributions
Methodology: Yating Zhu, Meifang Peng, Xiaoyan Wang; Formal analysis and investigation: Yating Zhu, Meifang Peng; Writing - original draft preparation: Yating Zhu, Meifang Peng; Writing - review and editing: Xiaoyan Wang; Weiwei Jiang, Ming Xia; Funding acquisition: Xiaoyan Wang, Xiaojie Huang, Weiwei Jiang, Ming Xia; Resources: Xiaojie Huang, Xiaoting Shen; Supervision: Xiaoyan Wang, Xiaojie Huang
Corresponding author
Ethics declarations
Conflict of Interest
The authors have no competing interests to declare that are relevant to the content of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix
Appendix
We train our model and all comparison models on two large datasets to further validate the effectiveness of our model. The PICCOLO-seg datasetFootnote 4 contains 3433 images from clinical colonoscopy videos, including white light and narrow band imaging (NBI) images. It includes 76 different lesions from 48 patients. We follow the official training and validation splits provided to train and evaluate models. The brain CA1 hippocampal region dataset is an electron microscopy image datasetFootnote 5 [62] (Hereinafter referred to as EM) for segmenting mitochondria. All images is taken from 5x5x5µm slices of the CA1 hippocampal region of the brain and contains 165 slices for both the training and test sets. Following the dataset split method in [63], we obtained 660 images for both the training set and the validation set. The results are shown in the Table 8. Overall, these models can draw similar conclusions on these two datasets as on the other previous used datasets. Specifically, the challenges in polyp segmentation mainly arise from the irregular shapes of the polyps and the unclear boundaries between the segmentation targets and the background, which require the model to have strong capabilities in detail feature extraction. As shown in Table 8, CNN-based models such as U-Net and CE-Net achieved better segmentation results compared to Transformer-based model Swin-Unet and models combining CNNs with Transformers like TransUNet. With the introduction of DACA blocks and SGA blocks, our model is able to simultaneously extract both global and local features, which helps capture multi-scale features for finer segmentation. As a result, our model achieves the best segmentation results in the PICCOLO-seg dataset. Although our model slightly underperforms U-Net in terms of accuracy, it outperforms U-Net by 7.94% and 9.15% in DSC and IoU, respectively. Similarly, in the EM dataset, mitochondria tend to be in close contact with other instances, which increases the difficulty of segmentation. On this dataset, LGCE-Net achieves the best segmentation results.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhu, Y., Peng, M., Wang, X. et al. LGCE-Net: a local and global contextual encoding network for effective and efficient medical image segmentation. Appl Intell 55, 66 (2025). https://doi.org/10.1007/s10489-024-05900-5
Accepted:
Published:
DOI: https://doi.org/10.1007/s10489-024-05900-5