Abstract
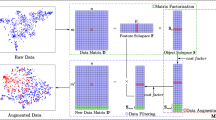
Disease diagnosis tasks using microbial data are often hindered by extreme class imbalance issues, which are further manifested as inter-class and intra-class imbalances. The former can be handled by general methods such as the SMOTE, while the latter has not been well studied. In this paper, we propose an ensemble classification algorithm based on space partitioning and data augmentation (ECSD) to address both types of imbalances. First, the data are mapped into a low-dimensional space through KPCA, LMNN, and RENN. These techniques address the data sparsity and noise in the original dataset. Second, we design a Kannoy technique to increase the distance between data points in different subspaces. In this way, the data distribution is more uniform, thus alleviating the intra-class imbalance problem. Third, a WGAN trained on the whole dataset is used to augment the data in each subspace. Different data augmentation and filtering strategies are employed to alleviate inter-class imbalance issues. Finally, base classifiers trained on each subspace are ensembled using a distance-weighted technique. The ensembler aims to provide stable predictions. Our algorithm is compared with four algorithms for handling class imbalance and three algorithms that address microbial-based diagnosis on 17 datasets. The results show that our algorithm outperforms its counterparts in terms of multiple metrics, especially when the dataset imbalance ratio is high.

















Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Data Availability
The data that support the findings of this study are available on github
References
Andoni A (2011) Nearest neighbor search in high-dimensional spaces. Math Found Comput 2011:1–1. https://doi.org/10.1007/978-3-642-22993-0_1
Anthony B, Nabil A, Bastien C (2021) Toward informed partitioning for load balancing: A proof-of-concept. J Comput Sci 61:101644. https://doi.org/10.1016/j.jocs.2022.101644
Armstrong G, Rahman G, Martino C, McDonald D, Gonzalez A, Mishne G, Knight R (2022) Applications and comparison of dimensionality reduction methods for microbiome data. Front Bioinform 2:821861. https://doi.org/10.3389/fbinf.2022.821861
Barua S, Islam MM, Yao X, Murase K (2014) Mwmote-majority weighted minority oversampling technique for imbalanced data set learning. IEEE Trans Knowl Data Eng 26(2):405–42. https://doi.org/10.1109/TKDE.2012.232
Bernhardsson E, et al. (2018) Annoy (approximate nearest neighbors oh yeah). https://github.com/spotify/annoy
Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP (2002) Smote: synthetic minority over-sampling technique. J Artif Intell Res 16:321–35. https://doi.org/10.1613/jair.953
Chen S, He H, Garcia EA (2010) Ramoboost: Ranked minority oversampling in boosting. IEEE Trans Neural Netw 21(10):1624–1642. https://doi.org/10.1109/TNN.2010.2066988
Chen ZL, Fu LL, Yao J, Guo WZ, Plant C, Wang SP (2023) Learnable graph convolutional network and feature fusion for multi-view learning. Inform Fusion 95:109–11. https://doi.org/10.1016/j.inffus.2023.02.013
Chris S, Taghi MK, Jason VH, Amri N (2010) Rusboost: A hybrid approach to alleviating class imbalance. IEEE Trans Syst Man Cybern Syst Hum 40(1):185–197. https://doi.org/10.1109/TSMCA.2009.2029559
Dang T, Kumaishi K, Usui E, Kobori S, Sato T, Toda Y, Yamasaki Y, Tsujimoto H, Ichihashi Y, Iwata H (2022) Stochastic variational variable selection for high-dimensional microbiome data. Microbiome 10(1):1–18. https://doi.org/10.1186/s40168-022-01439-0
Dekaboruah E, Suryavanshi MV, Chettri D, Verma AK (2020) Human microbiome: an academic update on human body site specific surveillance and its possible role. Arch Microbiol 202(8):2147–2167. https://doi.org/10.1007/s00203-020-01931-x
Dong M, Li Lh, Chen M, Kusalik A, Xu W (2020) Predictive analysis methods for human microbiome data with application to parkinson’s disease. PLoS ONE 15(8):e023777. https://doi.org/10.1371/journal.pone.0237779
Friedman JH (2001) Greedy function approximation: a gradient boosting machine. Ann Stat 29(5):1189–123. https://doi.org/10.1214/aos/1013203451
Georgios D, Fernando B, Felix L (2018) Improving imbalanced learning through a heuristic oversampling method based on k-means and smote. Inf Sci 465:1–2. https://doi.org/10.1016/j.ins.2018.06.056
Han H, Wang WY, Mao BH (2005) Borderline-smote: A new over-sampling method in imbalanced data sets learning. In: Advances in Knowledge Discovery and Data Mining, pp 878–88https://doi.org/10.1007/11538059_91
He GL, Zhao W, Xia XW, Peng R, Wu XY (2019) An ensemble of shapelet-based classifiers on inter-class and intra-class imbalanced multivariate time series at the early stage. Soft Comput 23(18):6097–611. https://doi.org/10.1007/s00500-018-3261-3
He HB, Bai Y, Edwardo A G, Li ST (2008) Adasyn: Adaptive synthetic sampling approach for imbalanced learning. In: 2008 IEEE International Joint Conference on Neural Networks (IEEE World Congress on Computational Intelligence). pp 1322–132. https://doi.org/10.1109/IJCNN.2008.4633969
Johnson JM, Khoshgoftaar TM (2021) The class imbalance problem. Nat Methods 18(11):1270–127. https://doi.org/10.1038/s41592-021-01302-4
Khushi M, Shaukat K, Talha Alam M, Hameed IA, Uddin S, Luo SH, Yang XY, Reyes MC (2021) A comparative performance analysis of data resampling methods on imbalance medical data. IEEE Access 9:109960–109975. https://doi.org/10.1109/ACCESS.2021.3102399
Li Q, Yang Y, Wu ZD, Ding T (2021) Review of gut microbiome analysis prediction models and algorithms. Microbiol China 48(1):180–196. https://doi.org/10.13344/j.microbiol.china.200346
Lin WC, Tsai CF, Hu YH, Jhang JS (2017) Clustering-based undersampling in class-imbalanced data. Inf Sci 409–410:17–26. https://doi.org/10.1016/j.ins.2017.05.008
Liu XY, Wu JX, Zhou ZH (2009) Exploratory undersampling for class-imbalance learning. IEEE Trans Syst Man Cybern B Cybern 39(2):539–55. https://doi.org/10.1109/TSMCB.2008.2007853
Liu ZN, Cao W, Gao ZF, Bian J, Chen HC, Chang Y, Liu TY (2020) Self-paced ensemble for highly imbalanced massive data classification. In: 2020 IEEE 36th International Conference on Data Engineering (ICDE). pp 841–852. https://doi.org/10.1109/ICDE48307.2020.00078
M NM (2002) Clustering large data sets. In: Soft computing approach to pattern recognition and image processing. pp 41–6. https://doi.org/10.1142/9789812776235_0003
Mohammed R, Rawashdeh J, Abdullah M (2020) Machine learning with oversampling and undersampling techniques: Overview study and experimental results. In: 2020 11th International Conference on Information and Communication Systems (ICICS). pp 243–2. https://doi.org/10.1109/ICICS49469.2020.239556
Pattaramon V, Eyad E, Andrei P (2021) On the class overlap problem in imbalanced data classification. Knowl-Based Syst 212:106631. https://doi.org/10.1016/j.knosys.2020.106631
Shorten C, Khoshgoftaar TM (2019) A survey on image data augmentation for deep learning. J Big Data 6(1):1–4. https://doi.org/10.1186/s40537-019-0197-0
Wang HY, Wang JS, Zhu LF (2021) A new validity function of fcm clustering algorithm based on intra-class compactness and inter-class separation. J Intell Fuzz Syst 40(6):12411–12432. https://doi.org/10.3233/JIFS-210555
Wang KF, Gou C, Duan YJ, Lin YL, Zheng XH, Wang FY (2017) Generative adversarial networks: introduction and outlook. IEEE/CAA J Automatica Sinica 4(4):588–59. https://doi.org/10.1109/JAS.2017.7510583
Wen LY, Chen Z, Xie XN, Min F (2023) Microbial data augmentation combining feature extraction and transformer network. Int J Mach Learn Cybern. https://doi.org/10.1007/s13042-023-02047-6
Wen LY, Wang X, Min F (2023) Cost-sensitive microbial data augmentation through matrix factorization. Appl Intell 12684–127. https://doi.org/10.1007/s10489-022-04187-8
Wen LY, Zhang XM, Li QF, Min F (2023) Kga: integrating kpca and gan for microbial data augmentation. Int J Mach Learn Cybern 1427–144. https://doi.org/10.1007/s13042-022-01707-3
Yang FL, Zou Q (2020) mAML: an automated machine learning pipeline with a microbiome repository for human disease classification. Database 2020. https://doi.org/10.1093/database/baaa050
Acknowledgements
This work was supported in part by the National Natural Science Foundation of China under grant Nos. 62136002 and 61876027.
Author information
Authors and Affiliations
Contributions
Liu-Ying Wen is responsible for conceptualization, methodology, writing, and Software. Zhu Chen is responsible for data preprocessing, and Writing. Fan Min is responsible for conceptualization, and writing-reviewing.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no financial and personal relationships with other people or organizations that can inappropriately influence our work. There is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the position presented in, or the review of, the manuscript entitled.
Ethical and Informed Consent
This paper does not contain any studies with animals performed by any of the authors. Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wen, LY., Chen, Z. & Min, F. Ensemble microbial classification based on space partitioning and data augmentation. Appl Intell 55, 47 (2025). https://doi.org/10.1007/s10489-024-05961-6
Accepted:
Published:
DOI: https://doi.org/10.1007/s10489-024-05961-6