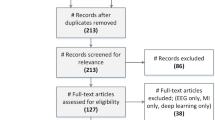
Abstract
The K-complex is one of the most important and noticeable features in the electroencephalography (EEG) signal, therefore its detection is critical for EEG signal analysis. It is used to diagnose neurophysiologic and cognitive disorders as well as sleep studies. Existing detection methods largely depend on tedious, time-consuming, and error-prone manual inspection of the EEG waveform. In this paper, a highly accurate K-complex detection system is developed. Based on multiple convolutional neural network (CNN) feature extraction backbones and EEG waveform images, you only look once v3 (YOLOv3) detector was designed, trained, and tested. Extensive performance evaluation was performed using five deep transfer learning feature extraction models; Darknet-53, MobileNets, ResNet-18, SqueezeNet, and Darknet-53-coco. The dataset was comprised of 10948 images of EEG waveforms, with the K-complex location automatically annotated with bounding boxes. The Darknet-53 model performed consistently high (i.e., 89.84–99.44% precision and 10.41–0.55% miss rate). Thus, it is possible to perform automatic K-complex detection in real-time with high accuracy that aid practitioners in speedy EEG inspection.











Similar content being viewed by others
Data availability
The dataset of EEG signals is publicly available from www.doi.org/10.5281/ZENODO.2650142. The images generated from the EEG signals during the current study are available from the corresponding author on reasonable request.
References
Cho, S.P., Lee, J., Park, H.D., Lee, K.J.: Detection of arousals in patients with respiratory sleep disorders using a single channel EEG. In: 2005 IEEE Engineering in Medicine and Biology 27th Annual Conference. IEEE, Shanghai (2005). https://doi.org/10.1109/iembs.2005.1617036
Smith, J.R., Funke, W.F., Yeo, W.C., Ambuehl, R.A.: Detection of human sleep EEG waveforms. Electroencephalogr. Clin. Neurophysiol. 38(4), 435–437 (1975). https://doi.org/10.1016/0013-4694(75)90269-2
Saccomandi, F., Priano, L., Mauro, A., Nerino, R., Guiot, C.: Automatic detection of transient EEG events during sleep can be improved using a multi-channel approach. Clin. Neurophysiol. 119(4), 959–967 (2008). https://doi.org/10.1016/j.clinph.2007.12.016
Hartmann, S., Baumert, M.: Automatic a-phase detection of cyclic alternating patterns in sleep using dynamic temporal information. IEEE Trans. Neural Syst. Rehabil. Eng. 27(9), 1695–1703 (2019). https://doi.org/10.1109/tnsre.2019.2934828
Sharma, M., Patel, V., Tiwari, J., Acharya, U.R.: Automated characterization of cyclic alternating pattern using wavelet-based features and ensemble learning techniques with EEG signals. Diagnostics 11(8), 1380 (2021). https://doi.org/10.3390/diagnostics11081380
Wen, D., Cheng, Z., Li, J., Zheng, X., Yao, W., Dong, X., Saripan, M.I., Li, X., Yin, S., Zhou, Y.: Classification of ERP signal from amnestic mild cognitive impairment with type 2 diabetes mellitus using single-scale multi-input convolution neural network. J. Neurosci. Methods 363, 109353 (2021). https://doi.org/10.1016/j.jneumeth.2021.109353
Wennberg, R.: Intracranial cortical localization of the human k-complex. Clin. Neurophysiol. 121(8), 1176–1186 (2010). https://doi.org/10.1016/j.clinph.2009.12.039
Gandhi, M.H., Emmady, P.D.: Physiology, k complex. StatPearls [Internet] (2021). Last accessed 15 March 2022
Bremer, G., Smith, J.R., Karacan, I.: Automatic detection of the k-complex in sleep electroencephalograms. IEEE Trans. Biomed. Eng. BME 17(4), 314–323 (1970). https://doi.org/10.1109/tbme.1970.4502759
Dumitrescu, C., Costea, I.-M., Cormos, A.-C., Semenescu, A.: Automatic detection of k-complexes using the cohen class recursiveness and reallocation method and deep neural networks with EEG signals. Sensors 21(21), 7230 (2021). https://doi.org/10.3390/s21217230
Al-Salman, W., Li, Y., Wen, P.: Detection of EEG k-complexes using fractal dimension of time frequency images technique coupled with undirected graph features. Front. Neuroinform. 13, 45 (2019). https://doi.org/10.3389/fninf.2019.00045
AL-Salman, W., Li, Y., Wen, P.: K-complexes detection in EEG signals using fractal and frequency features coupled with an ensemble classification model. Neuroscience 422, 119–133 (2019). https://doi.org/10.1016/j.neuroscience.2019.10.034
Kantar, T., Erdamar, A.: Detection of k-complexes in sleep EEG with support vector machines. In: 2017 25th Signal Processing and Communications Applications Conference (SIU), pp. 1–4 (2017). https://doi.org/10.1109/SIU.2017.7960311
Yücelbaş, C., Yücelbaş, Ş, Özşen, S., Tezel, G., Küççüktürk, S., Yosunkaya, Ş: A novel system for automatic detection of k-complexes in sleep EEG. Neural Comput. Appl. 29(8), 137–157 (2017). https://doi.org/10.1007/s00521-017-2865-3
Lajnef, T.: Meet spinky: an open-source spindle and k-complex detection toolbox validated on the open-access montreal archive of sleep studies (MASS). Front. Neuroinform. (2016). https://doi.org/10.3389/fninf.2017.00015
Patti, C.R., Abdullah, H., Shoji, Y., Hayley, A., Schilling, C., Schredl, M., Cvetkovic, D.: K-complex detection based on pattern matched wavelets. In: 2016 IEEE EMBS Conference on Biomedical Engineering and Sciences (IECBES). IEEE, Kuala Lumpur (2016). https://doi.org/10.1109/iecbes.2016.7843495
Lajnef, T., Chaibi, S., Eichenlaub, J.B., Ruby, P.M., Aguera, P.E., Samet, M., Kachouri, A., Jerbi, K.: Sleep spindle and k-complex detection using tunable q-factor wavelet transform and morphological component analysis. Front Hum Neurosci 9, 414 (2015). https://doi.org/10.3389/fnhum.2015.00414
Krohne, L.K., Hansen, R.B., Christensen, J.A.E., Sorensen, H.B.D., Jennum, P.: Detection of k-complexes based on the wavelet transform. In: 2014 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE, Chicago (2014). https://doi.org/10.1109/embc.2014.6944859
Zamir, Z.R., Sukhorukova, N., Amiel, H., Ugon, A., Philippe, C.: Optimization-based features extraction for k-complex detection. ANZIAM J 55, 384 (2014). https://doi.org/10.21914/anziamj.v55i0.7802
Zacharaki, E.I., Pippa, E., Koupparis, A., Kokkinos, V., Kostopoulos, G.K., Megalooikonomou, V.: One-class classification of temporal EEG patterns for k-complex extraction. In: 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC). IEEE, Osaka (2013). https://doi.org/10.1109/embc.2013.6610870
Shete, V.V., Sonar, S., Charantimatp, A., Elgendelwar, S.: Detection of k-complex in sleep EEG signal with matched filter and neural network. Int. J. Eng. Res. Technol. 4, 1–4 (2012)
Devuyst, S., Dutoit, T., Stenuit, P., Kerkhofs, M.: Automatic k-complexes detection in sleep EEG recordings using likelihood thresholds. In: 2010 Annual International Conference of the IEEE Engineering in Medicine and Biology. IEEE, Buenos Aires (2010). https://doi.org/10.1109/iembs.2010.5626447
Strungaru, C., Popescu, M.S.: Neural network for sleep EEG k-complex detection. Biomed. Tech. 43(s3), 113–116 (1998). https://doi.org/10.1515/bmte.1998.43.s3.113
...Gill, S.S., Xu, M., Ottaviani, C., Patros, P., Bahsoon, R., Shaghaghi, A., Golec, M., Stankovski, V., Wu, H., Abraham, A., Singh, M., Mehta, H., Ghosh, S.K., Baker, T., Parlikad, A.K., Lutfiyya, H., Kanhere, S.S., Sakellariou, R., Dustdar, S., Rana, O., Brandic, I., Uhlig, S.: AI for next generation computing: emerging trends and future directions. Internet Things 19, 100514 (2022). https://doi.org/10.1016/j.iot.2022.100514
Yu, Z., Wang, K., Wan, Z., Xie, S., Lv, Z.: Popular deep learning algorithms for disease prediction: a review. Clust. Comput. (2022). https://doi.org/10.1007/s10586-022-03707-y
Kumar, M.R., Rao, Y.S.: Epileptic seizures classification in EEG signal based on semantic features and variational mode decomposition. Clust. Comput. 22(S6), 13521–13531 (2018). https://doi.org/10.1007/s10586-018-1995-4
Devuyst, S.: The DREAMS Databases and Assessment Algorithm. Zenodo (2005) https://doi.org/10.5281/ZENODO.2650142. zenodo.org/record/2650142
Redmon, J., Farhadi, A.: YOLOv3: An Incremental Improvement (2018). http://arxiv.org/abs/1804.02767
Deng, J., Dong, W., Socher, R., Li, L.-J., Li, K., Fei-Fei, L.: Imagenet: A large-scale hierarchical image database. In: 2009 IEEE Conference on Computer Vision and Pattern Recognition, pp. 248–255 (2009). https://doi.org/10.1109/CVPR.2009.5206848
Redmon, J.: Darknet: Open Source Neural Networks in C (2013–2016). http://pjreddie.com/darknet/
Sandler, M., Howard, A., Zhu, M., Zhmoginov, A., Chen, L.-C.: Mobilenetv2: Inverted residuals and linear bottlenecks. In: 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 4510–4520 (2018). https://doi.org/10.1109/CVPR.2018.00474
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 770–778 (2016). https://doi.org/10.1109/CVPR.2016.90
Iandola, F.N., Moskewicz, M.W., Ashraf, K., Han, S., Dally, W.J., Keutzer, K.: Squeezenet: Alexnet-level accuracy with 50x fewer parameters and<1mb model size. arXiv (2016)
Lin, T.-Y., Maire, M., Belongie, S., Bourdev, L., Girshick, R., Hays, J., Perona, P., Ramanan, D., Zitnick, C.L., Dollár, P.: Microsoft COCO: Common Objects in Context. arXiv (2014). https://doi.org/10.48550/ARXIV.1405.0312. https://arxiv.org/abs/1405.0312
Sarker, I.H.: Deep learning: a comprehensive overview on techniques, taxonomy, applications and research directions. SN Comput. Sci. 2(6), 1–20 (2021). https://doi.org/10.1007/s42979-021-00815-1
Al-Salman, W., Li, Y., Wen, P.: Detection of k-complexes in EEG signals using a multi-domain feature extraction coupled with a least square support vector machine classifier. Neurosci. Res. 172, 26–40 (2021). https://doi.org/10.1016/j.neures.2021.03.012
Oliveira, G.H.B.S., Coutinho, L.R., da Silva, J.C., Pinto, I.J.P., Ferreira, J.M.S., Silva, F.J.S., Santos, D.V., Teles, A.S.: Multitaper-based method for automatic k-complex detection in human sleep EEG. Expert Syst. Appl. 151, 113331 (2020). https://doi.org/10.1016/j.eswa.2020.113331
Ranjan, R., Arya, R., Fernandes, S.L., Sravya, E., Jain, V.: A fuzzy neural network approach for automatic k-complex detection in sleep EEG signal. Pattern Recognit. Lett. 115, 74–83 (2018). https://doi.org/10.1016/j.patrec.2018.01.001
Ghanbari, Z., Moradi, M.: K-complex detection based on synchrosqueezing transform. AUT J. Electr. Eng. 49(2), 214–222 (2017). https://doi.org/10.22060/eej.2017.12577.5096
Patti, C.R., Abdullah, H., Shoji, Y., Hayley, A., Schilling, C., Schredl, M., Cvetkovic, D.: K-complex detection based on pattern matched wavelets. In: 2016 IEEE EMBS Conference on Biomedical Engineering and Sciences (IECBES), pp. 470–474 (2016). https://doi.org/10.1109/IECBES.2016.7843495
Acknowledgements
This work would not be possible without the financial support of Jordan University of Science and Technology, deanship of research.
Funding
This work was funded by Jordan University of Science and Technology (JUST), deanship of research, award number 20220146.
Author information
Authors and Affiliations
Contributions
Conceptualization: NK, and LF; Methodology: NK, and MF; Formal analysis and investigation: NK, and MF; Writing-original draft preparation: MF; Writing-review and editing: NK, MF, and LF; Funding acquisition: MF and LF; Resources: MF; Supervision: NK, and LF. Visualization: NK
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical statement
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Khasawneh, N., Fraiwan, M. & Fraiwan, L. Detection of K-complexes in EEG signals using deep transfer learning and YOLOv3. Cluster Comput 26, 3985–3995 (2023). https://doi.org/10.1007/s10586-022-03802-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10586-022-03802-0