Abstract
CGP-Library is an open-source, cross-platform written in C which implements Cartesian Genetic Programming (CGP) and its variations. It solves both academic and real-world problems. Since its inicial release, it has undergone some refinements, and it is without doubt the best supported toolkit for CGP. In this article it is presented a critical assessment of the CGP-Library features, its main strengths and its weaknesses.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
CGP-Library: http://www.cgplibrary.co.uk.
CGP-Library is an open-source, cross-platform library written in C which implements Cartesian Genetic Programming (CGP), Recurrent Cartesian Genetic Programming, Cartesian Genetic Programming of Artificial Neural Networks and Recurrent Cartesian Genetic Programming of Artificial Neural Networks. It solves both academic and real-world problems.
2 Background
The development of CGP-Library began in 2014. In 2015 a letter presenting it was published in Genetic Programming and Evolvable Machines [1]. Since its initial release, it has undergone some refinements, such as adding more functions, multiple thread support, creating cyclic programs and bug fixes. It is now at version 2.4. CGP-Library is without doubt the best supported toolkit for Cartesian Genetic Programming.
CGP-Library is maintained by Andrew Turner and can also be downloaded from https://github.com/andrewjamesturner/cgp-library.
3 Major features
CGP-Library can be run on Linux, Microsoft Windows and Apple Mac operating systems. It is designed to be used in teaching, academic research and real-world applications. Its application programming interface (API) provides functions for high level applications, customized algorithms and allows for the use of the generated results.
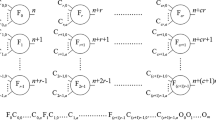
Each evolved chromosome is a graph composed of data flow links and nodes. There are 30 types of nodes, which are divided into four main categories: mathematical operations, logic gates, neuron transfer/activation functions and some “others” (that generate constants, random numbers, etc.).
4 Strengths
CGP-Library has undergone several updates in the three years since its initial version. These include new types of mutation, most of the benchmark problems and support for Recurrent Neural Networks. The latest changes include support for multi-thread evaluation via OpenMP.
In many cases there is no need to understand the details of the library’s own code or to edit it. Its API is very well documented. Thus any programmer even with just a basic programming background, can easily use the library for the most diverse of applications. The CGP-Library software package comes with twelve tutorials and a number of examples that show, step by step, the process of installing the library and setting it up to solve an example symbolic regression problem. The tutorials continue by showing how to customize the CGP graph nodes, the fitness function, the selection and reproduction schemes, the manipulation of data sets and chromosomes and how to visualise them. The tutorials explain how to use the command prompt to achieve this. They then continue to show how to store results using CSV files, the plotting of graphs and the generation of statistical reports. Finally they cover more advanced topics such as the evolution of neural networks and the use of multiple OpenMP threads.
All this can be achieved through the API. It is only necessary to edit one of the supplied example templates. This makes it the most intuitive and flexible library for using CGP that we have seen. Also, because it is open source and very well structured, more advanced users can readily add new features to it, or even modify existing ones. For example, a new customized mutation method could be added. In addition to all the above advantages, it can be compiled for a wide range of operating systems, and, as the library is compiled, it can be imported to Python, for example, through the C-type modules of its standard library. Since there are specific functions in the API for handling chromosomes, such as loading, saving, optimizing and executing them, the evolved results can be distributed and exploited by other tools. Thus, for example, we imagine that an electrical engineer could use an evolved chromosome representing a digital circuit together with a synthesis tool to automatically program a Field Programmable Gate Array (FPGAs).
5 Weaknesses
Perhaps the biggest weakness is that the package does not have a graphical interface. It would be nice if lay users could use a GUI to choose genetic parameters, and function types. Or to import training tables or to visualise forms. Other possibilities include customizing the fitness function, the selection and reproduction schemes, manipulating the chromosomes including controlling the various forms of optimizing them. This would be a more intuitive and simple alternative that would certainly make the use of the library more popular.
Although the CGP Library is very simple and intuitive for users with some programming knowledge, it does not have a users’ discussion forum. This could be extremely relevant to them and also help when extending or supporting the library.
Finally, there is no support for generating results in hardware description languages (HDLs), e.g. for the synthesis of circuits to be embedded into FPGAs. Although it is possible to create suitable FPGA sequential logic from CGP Library’s output using external tools, it is not easy to see how it can be applied to circuits which are required to use state machines. Perhaps this might be solvable with some effort by inserting flip-flops as a new type of CGP node.
6 Conclusions
If you ask us which is the best toolkit for working with CGP, we would, without hesitation, recommend the CGP-Library. Less experienced users perhaps might prefer other simpler alternatives like the Java CGP Library. However, if you are a programmer or researcher the CGP-Library is unbeatable at the moment in terms of its characteristics, flexibility, extensibility and documentation.
Reference
A.J. Turner, J.F. Miller, Introducing a cross platform open source cartesian genetic programming library. J Genet Program Evolvable Mach 16, 83–91 (2014)
Acknowledgements
We thank FAPESP for financial support under Grant 2015/23297-4.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Pedrino, E.C., Paris, P.C.D., de Lima, D.P. et al. Software review: CGP-Library. Genet Program Evolvable Mach 18, 279–281 (2017). https://doi.org/10.1007/s10710-017-9303-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10710-017-9303-2