Abstract
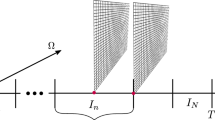
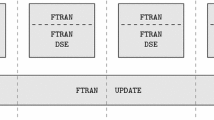
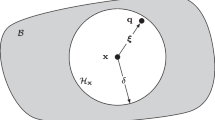
Cardiovascular diseases are associated with high mortality rates in the globe. The development of new drugs, new medical equipment and non-invasive techniques for the heart demand multidisciplinary efforts towards the characterization of cardiac anatomy and function from the molecular to the organ level. Computational modeling has demonstrated to be a useful tool for the investigation and comprehension of the complex biophysical processes that underlie cardiac function. The set of Bidomain equations is currently one of the most complete mathematical models for simulating the electrical activity in cardiac tissue. Unfortunately, large scale simulations, such as those resulting from the discretization of an entire heart, remain a computational challenge. In order to reduce simulation execution times, parallel implementations have traditionally exploited data parallelism via numerical schemes based on domain-decomposition. However, it has been verified that the parallel efficiency of these implementations severely degrades as the number of processors increases. In this work we propose and implement a new parallel algorithm for the solution of cardiac models. By relaxing the coherence of the execution, a new level of parallelism could be identified and exploited: pipelining. A synchronous parallel algorithm that uses both pipelining and data decomposition techniques was implemented and used the MPI library for communication. Numerical tests were performed in two different cluster configurations. Our preliminary results indicated that the proposed algorithm is able to increase the parallel efficiency up to 20% on an 8-core cluster. On a 32-core cluster the multi-level algorithm was 1.7 times faster than the traditional domain decomposition algorithm. In addition, the numerical precision was kept under control (relative errors under 6%) when the relaxed coherence execution was adopted.
Similar content being viewed by others
References
Hodgkin A. L., Huxley A. F.: A quantitative description of membrane current and its application to conduction and excitation in nerve. J. Physiol. 117, 500–544 (1952)
Plonsey, R.: Bioelectric sources arising in excitable fibers (ALZA lecture). Ann. Biomed. Eng. 16(6), 519–546 (1988). [Online]. Available: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?cmd=prlinks&dbfrom=pubmed&retmode=ref&id=3067629
Gima K., Rudy Y.: Ionic current basis of electrocardiographic waveforms: A model study. Circ. Res. 90, 889–896 (2002)
Santos R., Olaf K., Steinhoff U., Bauer S., Trahms L., Koch H.: Mcg to ecg source differences: measurements and a 2d computer model study. J. Electrocardiol. 37(4), 123–127 (2004)
Vigmond E., Aguel F., Trayanova N.: Computational techniques for solving the bidomain equations in three dimensions. IEEE Trans. Biomed. Eng. 49(11), 1260–1269 (2002)
Sundnes, J., Lines, G. T., Tveito, A.: An operator splitting method for solving the bidomain equations coupled to a volume conductor model for the torso. Math. Biosci. 194(2), 233–248 (2005). [Online]. Available: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?cmd=prlinks&dbfrom=pubmed&retmode=ref&id=15854678
Weber dos Santos, R., Plank, G., Bauer, S., Vigmond, E.: Parallel multigrid preconditioner for the cardiac bidomain model. IEEE Trans. Biomed. Eng. 51(11), 1960–1968 (2004). [Online]. Available: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?cmd=prlinks&dbfrom=pubmed&retmode=ref&id=15536898
Plank G., Liebmann M., Weber dos Santos R., Vigmond E., Haase G.: Algebraic Multigrid Preconditioner for the Cardiac Bidomain Model. IEEE Trans. Biomed. Eng. 54, 585–596 (2007)
Felippa C. A., Park K. C., Farhat C.: Partitioned analysis of coupled mechanical systems. Comput. Methods Appl. Mech. Eng. 190, 3247–3270 (2001)
Henriquez C.: Simulating the electrical behavior of cardiac tissue using the bidomain model. Crit. Rev. Biomed. Eng. 21(1), 1–77 (1993)
Strikwerda, J.: Finite difference schemes and partial differential equations. Soc. Ind. Math. (2004)
Message Passing Interface Forum: MPI, a message-passing interface standard. Int. J. Supercomp. 8, 159–416 (1994)
Balay, S., Buschelman, K., Gropp, W.D., Kaushik, D., Knepley, M., McInnes, L.C., Smith, B.F., Zhang, H.: PETSc users manual, Argonne National Laboratory, Tech. Rep. ANL-95/11 - Revision 2.1.5 (2002)
Santos R., Campos F., Neumann L., Nygren A., Giles W.: ATX-II effects on the apparent location of m cells in a computational human left ventricular wedge. J. Cardiovasc. Electrophysiol. 17, S86–S95 (2006)
Bauer, S., dos Santos, R., Schmal, T., Nagel, E., Baer, M., Koch, H.: QRS width and QT time alteration due to geometry change in modelled human cardiac magnetograms. In: Proceedings of IEEE Computers in Cardiology, pp. 639–642 (2005)
Muzikant A., Henriquez C.: Validation of three-dimensional conduction models using experimental mapping: are we getting closer?. Prog. Biophys. Mol. Biol. 69(2–3), 205–223 (1998)
Ten Tusscher K., Noble D., Noble P. J., Panfilov A. V.: A model for human ventricular tissue. Am. J. Physiol. Heart Circ. Physiol. 286(4), 1573–1589 (2004)
Skouibine, K., Trayanova, N., Moore, P.: A numerically efficient model for simulation of defibrillation in an active bidomain sheet of myocardium. Math. Biosci. 166(1), 85–100 (2000) [Online]. Available: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?cmd=prlinks&dbfrom=pubmed&retmode=ref&id=10882801
Roth B.: Meandering of spiral waves in anisotropic cardiac tissue. Physica D 150, 127–136 (2001)
Jacquemet, V., Henriquez, C.: Finite volume stiffness matrix for solving anisotropic cardiac propagation in 2-D and 3-D unstructured meshes. IEEE Trans. Biomed. Eng., 52(8), 1490–1492 (2005). [Online]. Available: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?cmd=prlinks&dbfrom=pubmed&retmode=ref&id=16119246
Plank, G., Leon, L. J., Kimber, S., Vigmond, E. J.: Defibrillation depends on conductivity fluctuations and the degree of disorganization in reentry patterns. J. Cardiovasc. Electrophysiol. 16(2), 205–216 (2005). [Online]. Available: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?cmd=prlinks&dbfrom=pubmed&retmode=ref&id=15720461
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Xavier, C.R., Oliveira, R.S., da Fonseca Vieira, V. et al. Multi-Level Parallelism for the Cardiac Bidomain Equations. Int J Parallel Prog 37, 572–592 (2009). https://doi.org/10.1007/s10766-009-0110-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10766-009-0110-0