Abstract
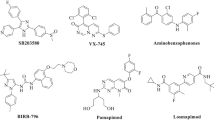
The p38 protein kinase is a serine–threonine mitogen activated protein kinase, which plays an important role in inflammation and arthritis. A combined study of 3D-QSAR and molecular docking has been undertaken to explore the structural insights of pyrazolyl urea p38 kinase inhibitors. The 3D-QSAR studies involved comparative molecular field analysis (CoMFA) and comparative molecular similarity indices (CoMSIA). The best CoMFA model was derived from the atom fit alignment with a cross-validated r 2 (q 2) value of 0.516 and conventional r 2 of 0.950, while the best CoMSIA model yielded a q 2 of 0.455 and r 2 of 0.979 (39 molecules in training set, 9 molecules in test set). The CoMFA and CoMSIA contour maps generated from these models provided inklings about the influence of interactive molecular fields in the space on the activity. GOLD, Sybyl (FlexX) and AutoDock docking protocols were exercised to explore the protein–inhibitor interactions. The integration of 3D-QSAR and molecular docking has proffered essential structural features of pyrazolyl urea inhibitors and also strategies to design new potent analogues with enhanced activity.









Similar content being viewed by others
References
Pearson G, Robinson F, Beers T, Xu E, Berman K, Cobb M (2001) Endocr Rev 22:153
Kulkarni RG, Achaiah G, Sastry GN (2006) Curr Pharm Des 12:2437
Ono K, Han J (2000) Cell Sign 12:1
Zarubin T, Han J (2005) Cell Res 15:11
Schaeffer H, Weber M (1999) Mol Cell Biol 19:2435
English J, Pearson G, Wilsbacher J, Swantek J, Karandikar M, Xu S, Cobb MH (1999) Exp Cell Res 253:255
Behr TM, Berova M, Doe CP, Ju H, Angermann CE, Boehm J, Willette RN (2003) Cur Opin Invest Drugs 4:1059
Jiang Y, Chen C, Li Z, Guo W, Gegner A, Lins S, Han J (1996) J Biol Chem 271:17920
Li Z, Jiang Y, Ulevitch J, Han J (1996) Biochem Biophys Res Comm 228:334
Jiang Y, Gram H, Zhao M, New L, Gu J, Feng L, Padova F, Ulevitch R, Han J (1997) J Biol Chem 272:30122
Hanks SK, Hunter T (1999) FASEB J 9:576
New L, Jiang Y, Zhao M, Liu K, Zhu W, Flood LJ, Kato Y, Parry G, Han J (1998) EMBO J 17:3372
Newton R, Holden N (2003) Biodrugs 17:113
Jackson R, Bolognese B, Hillegass L, Kassis S, Adams J, Griswold DE, Winkler JD (1998) J Pharmacol Exp Ther 284:687
Lee JC, Kumar S, Griswold DE, Underwood DC, Votta BJ, Adams JL (2000) Immunopharmacology 47:185
Lee C, Laydon J, McDonnell P, Gallagher TF, Kumar S, Green D, McNulty D, Blumenthal MJ, Heys JR, Landvatter SW (1994) Nature 372:739
Gallagher TF, Susan M, Thomson SM, Garigipati RS, Sorenson ME, Smietana JM, Lee D, Bender PE, Lee JC, Laydon JT, Griswold DE (1995) Bioorg Med Chem Let 5:1171
Wang Z, Canagarajah J, Boehm JC, Kassisa S, Cobb MH, Young PR, Abdel-Meguid S, Adams JL, Goldsmith EJ (1998) Structure 6:1117
Young PR, McLaughlin MM, Kumar S, Kasis S, Doyle ML, McNultry D, Gallagher TF, Fisher S, McDonnell PC, Carr SA, Huddleston MJ, Seibel G, Porter TG, Livi GP, Adams JL, Lee JC (1997) J Biol Chem 272:12116
Laufer S, Wagner G, Kotschenreuther A, Albrecht W (2003) J Med Chem 46:3230
Cirillo F, Pargellis C, Regan J (2002) Curr Top Med Chem 2:1021
Stelmach E, Liu L, Patel B, Pivnichny V, Scapin G, Singh B Hop CECA, Wang Z, Strauss JR, Cameron PM, Nichols EA, O’Keefe SJ, O’Neill EA, Schmatz DM, Schwartz CD, Thompson CM, Dennis M, Zaller DM, Doherty JB (2003) Bioorg Med Chem Lett 13:277
Lee M, Dominguez C (2005) Cur Med Chem 12:2979
Dumas J, Hatoum-Mokda H, Siblety R, Riedl B, Scott J, Monahan MK, Lowinger TB, Brennan C, Natero R, Turner T, Johnson JS, Schoenleber R, Bhargava A, Wilhelm SM, Housley TJ, Ranges GE, Shrikhande A (2000) Bioorg Med Chem Letts 10:2051
Dumas J, Hatoum-Mokda H, Siblety R, Riedl B, Scott J, Khire U, Lee W, Wood J, Wolanin D, Cooley J, Bankston D, Redman AM, Schoenleber R, Caringal Y, Gunn D, Romero R, Osterhout M, Paulsen H, Housley TJ, Wilhelm SM, Pirro J, Chien D, Ranges GE, Shrikhande A, Muzsi A, Bortolon E, Wakefield J, Ostravage CG, Bhargava A, Chaub T (2002) Bioorg Med Chem Letts 12:1559
Regan J, Pargellis CA, Cirillo PF, Gilmore T, Hickey ER, Peet GW, Proto A, Swinamer A, Moss N (2003) Bioorg Med Chem Letts 13:3101
Pargellis C, Tong L, Churchill L, Cirrillo F, Gilmore T, Graham AG, Grob PM, Hickey ER, Moss N, Pav S, Regan J (2002) Nat Struct Biol 9:268
Cramer D, Patterson E, Bunce D (1988) J Am Chem Soc 110:5959
Klebe G, Abraham U, Mietzner T (1994) J Med Chem 37:4130
Klebe G, Abraham U (1999) J Comp Aided Mol Des 13:1
Sybyl 6.9, Tripos Inc., 1699 S. Hanley Rd., St Leuis, MO 63144 USA
Viswanadhan VN, Ghose AK, Revankar GR, Robins RK (1989) J Chem Inf Comput Sci 29:163
Geladi P (1998) J Chemon 2:231
Cramer RD, Bunce D, Patterson E (1988) Quant Struct Act Relat 7:18
GOLD, version 2.2, Cambridge Crystallographic Data Centre: Cambridge, U.K
Rarey M, Kramer B, Lengauer T, Klebe G (1996) J Mol Biol 261:470
Kramer B, Rarey M, Lengauer T (1999) Proteins: Str Func Genet 37:228
Goodsell DS, Olson AJ (1990) Proteins: Str Func Genet 8:195
Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ (1998) J Comput Chem 19:1639
Acknowledgements
RG thanks AICTE for providing the financial assistance and PS thanks CSIR for a senior research fellowship. The Director of IICT is thanked for his continued encouragement.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kulkarni, R.G., Srivani, P., Achaiah, G. et al. Strategies to design pyrazolyl urea derivatives for p38 kinase inhibition: a molecular modeling study. J Comput Aided Mol Des 21, 155–166 (2007). https://doi.org/10.1007/s10822-006-9092-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-006-9092-9