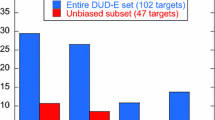
Abstract
Ligand enrichment among top-ranking hits is a key metric of virtual screening. To avoid bias, decoys should resemble ligands physically, so that enrichment is not attributable to simple differences of gross features. We therefore created a directory of useful decoys (DUD) by selecting decoys that resembled annotated ligands physically but not topologically to benchmark docking performance. DUD has 2950 annotated ligands and 95,316 property-matched decoys for 40 targets. It is by far the largest and most comprehensive public data set for benchmarking virtual screening programs that I am aware of. This paper outlines several ways that DUD can be improved to provide better telemetry to investigators seeking to understand both the strengths and the weaknesses of current docking methods. I also highlight several pitfalls for the unwary: a risk of over-optimization, questions about chemical space, and the proper scope for using DUD. Careful attention to both the composition of benchmarks and how they are used is essential to avoid being misled by overfitting and bias.



Similar content being viewed by others
References
Verdonk ML, Cole JC, Hartshorn MJ, Murray CW, Taylor RD (2003) Improved protein-ligand docking using GOLD. Proteins 52:609–623
Abagyan RA, Totrov MM, Kuznetsov DA (1994) ICM: a new method for structure modeling and design. J Comput Chem 14:488–506
Meng EC, Shoichet BK, Kuntz ID (1992) Automated docking with grid-based energy evaluation. J Comput Chem 13:505–524
McGann MR, Almond HR, Nicholls A, Grant JA, Brown FK (2003) Gaussian docking functions. Biopolymers 68:76–90
Friesner RA et al (2004) Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem 47:1739–1749
Rarey M, Kramer B, Lengauer T, Klebe G (1996) A fast flexible docking method using an incremental construction algorithm. J Mol Biol 261:470–489
Miller MD, Kearsley SK, Underwood DJ, Sheridan RP (1994) FLOG: a system to select ‘quasi-flexible’ ligands complementary to a receptor of known three-dimensional structure. J Comput Aided Mol Des 8:153–174
Perola E, Walters WP, Charifson PS (2004) A detailed comparison of current docking and scoring methods on systems of pharmaceutical relevance. Proteins 56:235–249
Ferrara P, Gohlke H, Price DJ, Klebe G, Brooks CL III (2004) Assessing scoring functions for protein-ligand interactions. J Med Chem 47:3032–3047
Kellenberger E, Rodrigo J, Muller P, Rognan D (2004) Comparative evaluation of eight docking tools for docking and virtual screening accuracy. Proteins 57:225–242
Kontoyianni M, McClellan LM, Sokol GS (2004) Evaluation of docking performance: comparative data on docking algorithms. J Med Chem 47:558–565
Wang R, Lu Y, Fang X, Wang S (2004) An extensive test of 14 scoring functions using the PDBbind refined set of 800 protein-ligand complexes. J Chem Inf Comput Sci 44:2114–2125
Verdonk ML et al (2004) Virtual screening using protein-ligand docking: avoiding artificial enrichment. J Chem Inf Comput Sci 44:793–806
Xing L, Hodgkin E, Liu Q, Sedlock D (2004) Evaluation and application of multiple scoring functions for a virtual screening experiment. J Comput Aided Mol Des 18:333–344
Onodera K, Satou K, Hirota H (2007) Evaluations of molecular docking programs for virtual screening. J Chem Inf Model 47:1609–1618
Zhou Z, Felts AK, Friesner RA, Levy RM (2007) Comparative performance of several flexible docking programs and scoring functions: enrichment studies for a diverse set of pharmaceutically relevant targets. J Chem Inf Model 47:1599–1608
Hartshorn MJ et al (2007) Diverse, high-quality test set for the validation of protein-ligand docking performance. J Med Chem 50:726–741
Nissink JW et al (2002) A new test set for validating predictions of protein-ligand interaction. Proteins 49:457–471
Kuntz ID, Chen K, Sharp KA, Kollman PA (1999) The maximal affinity of ligands. Proc Natl Acad Sci USA 96:9997–10002
Pham TA, Jain AN (2006) Parameter estimation for scoring protein–ligand interactions using negative training data. J Med Chem 49:5856–5868
Bissantz C, Folkers G, Rognan D (2000) Protein-based virtual screening of chemical databases: 1. evaluation of different docking/scoring combinations. J Med Chem 43:4759–4767
Irwin JJ, Shoichet BK (2005) ZINC–a free database of commercially available compounds for virtual screening. J Chem Inf Model 45:177–182
Halgren TA et al (2004) Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J Med Chem 47:1750–1759
Gohlke H, Hendlich M, Klebe G (2000) Knowledge-based scoring function to predict protein-ligand interactions. J Mol Biol 295:337–356
Ferrari AM, Wei BQ, Costantino L, Shoichet BK (2004) Soft docking and multiple receptor conformations in virtual screening. J Med Chem 47:5076–5084
van Drie JH (2003) Pharmacophore discovery–lessons learned. Curr Pharm Des 9:1649–1664
Brünger A (1992) The free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355:472–474
Byvatov E, Schneider G (2003) Support vector machine applications in bioinformatics. Appl Bioinformatics 2:67–77
Kleywegt GJ (2007) Separating model optimization and model validation in statistical cross-validation as applied to crystallography. Acta Crystallogr D Biol Crystallogr 63:939–940
Graves AP, Brenk R, Shoichet BK (2005) Decoys for docking. J Med Chem 48:3714–3728
Fink T, Reymond JL (2007) Virtual exploration of the chemical universe up to 11 atoms of C, N, O, F: assembly of 26.4 million structures (110.9 million stereoisomers) and analysis for new ring systems, stereochemistry, physicochemical properties, compound classes, and drug discovery. J Chem Inf Model 47:342–353
Hann MM, Oprea TI (2004) Pursuing the leadlikeness concept in pharmaceutical research. Curr Opin Chem Biol 8:255–263
James CA (2007) Daylight Theory Manual 4.93
Overington JP, Al-Lazikani B, Hopkins AL (2006) How many drug targets are there? Nat Rev Drug Discov 5:993–996
Cherezov V et al (2007) High-Resolution Crystal Structure of an Engineered Human {beta}2-Adrenergic G Protein Coupled Receptor. Science 366
Yohannan S, Hu Y, Zhou Y (2007) Crystallographic study of the tetrabutylammonium block to the KcsA K+ channel. J Mol Biol 366:806–814
Xiong JP et al (2002) Crystal structure of the extracellular segment of integrin alpha Vbeta3 in complex with an Arg-Gly-Asp ligand. Science 296:151–155
Berman HM et al (2000) The protein data bank. Nucl Acid Res 28:235–242
Benson ML, Smith RD, Khazanov NA, Dimcheff B, Beaver J, Dresslar P, Nerothin J, Carlson HA (2008) Binding MOAD, a high-quality protein-ligand database. NAR 36:D674–D678
Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK (2007) BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res 35:D198–201
Zhang J et al (2004) Development of KiBank, a database supporting structure-based drug design. Comput Biol Chem 28:401–407
Good AC, Oprea TI (2008) Optimization of CAMD techniques 3. Virtual screening enrichment studies: a help or hindrance in tool selection? J Comput Aided Mol Des this issue, doi:10.1007/s10822-007-9167-2
Acknowledgements
Supported by NIH grant GM71896 (to Brian K. Shoichet and J.J.I.). I thank Prof. Brian K. Shoichet for comments and suggestions arising from an ongoing discussion of this topic, and Dr. Peter Kolb, Kristin Coan and Michael Mysinger for reading the manuscript. I thank the reviewers for thoughtful and helpful suggestions.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Irwin, J.J. Community benchmarks for virtual screening. J Comput Aided Mol Des 22, 193–199 (2008). https://doi.org/10.1007/s10822-008-9189-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-008-9189-4