Abstract
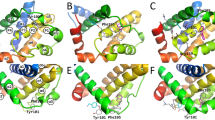
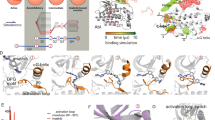
Detailed understanding of protein–ligand interactions is crucial to the design of more effective drugs. This is particularly true when targets are protein interfaces which have flexible, shallow binding sites that exhibit substantial structural rearrangement upon ligand binding. In this study, we use molecular dynamics simulations and free energy calculations to explore the role of ligand-induced conformational changes in modulating the activity of three generations of Bcl-XL inhibitors. We show that the improvement in the binding affinity of each successive ligand design is directly related to a unique and measurable reduction in local flexibility of specific regions of the binding groove, accompanied by the corresponding changes in the secondary structure of the protein. Dynamic analysis of ligand–protein interactions reveals that the latter evolve with each new design consistent with the observed increase in protein stability, and correlate well with the measured binding affinities. Moreover, our free energy calculations predict binding affinities which are in qualitative agreement with experiment, and indicate that hydrogen bonding to Asn100 could play a prominent role in stabilizing the bound conformations of latter generation ligands, which has not been recognized previously. Overall our results suggest that molecular dynamics simulations provide important information on the dynamics of ligand–protein interactions that can be useful in guiding the design of small-molecule inhibitors of protein interfaces.











Similar content being viewed by others
References
Sharma SK, Ramsey TM, Bair KW (2002) Curr Med Chem. Anti-Cancer Agents 2:311–330. doi:10.2174/1568011023354191
Arkin MR, Wells JA (2004) Nat Rev Drug Discov 3:301–317. doi:10.1038/nrd1343
Berg T (2003) Angew Chem Int Ed 42:2462–2481. doi:10.1002/anie.200200558
Yin H, Hamilton AD (2005) Angew Chem Int Ed 44:4130–4163. doi:10.1002/anie.200461786
Muchmore SW, Sattler M, Liang H et al (1996) Nature 381:335–341. doi:10.1038/381335a0
Oltersdorf T, Elmore SW, Shoemaker AR et al (2005) Nature 435:677–681. doi:10.1038/nature03579
Borner C (2003) Mol Immunol 39:615–647. doi:10.1016/S0161-5890(02)00252-3
Cory S, Adams JM (2002) Nat Rev Cancer 2:647–656. doi:10.1038/nrc883
van Delft MF, Huang DC (2006) Cell Res 16:203–213. doi:10.1038/sj.cr.7310028
Coultas L, Strasser A (2003) Semin Cancer Biol 13:115–123. doi:10.1016/S1044-579X(02)00129-3
Kirkin V, Joos S, Zornig M (2004) BBA-Mol Cell Res 1644:229–249
Amundson SA, Myers TG, Scudiero D et al (2000) Cancer Res 60:6101–6110
Green DR, Evan GI (2002) Cancer Cell 1:19–30. doi:10.1016/S1535-6108(02)00024-7
Wendt MD, Shen W, Kunzer A et al (2006) J Med Chem 49:1165–1181. doi:10.1021/jm050754u
Bruncko M, Oost TK, Belli BA et al (2007) J Med Chem 50:641–662. doi:10.1021/jm061152t
Sattler M, Liang H, Nettesheim D et al (1997) Science 275:983–986. doi:10.1126/science.275.5302.983
Petros AM, Nettesheim DG, Wang Y et al (2000) Protein Sci 9:2528–2534. doi:10.1017/S096183680000331X
Petros AM, Olejniczak ET, Fesik SW (2004) BBA-Mol Cell Res 1644:83–94
Shuker SB, Hajduk PJ, Meadows RP et al (1996) Science 274:1531–1534. doi:10.1126/science.274.5292.1531
Petros AM, Dinges J, Augeri DJ et al (2006) J Med Chem 49:656–663. doi:10.1021/jm0507532
Park CM, Oie T, Petros AM et al (2006) J Am Chem Soc 128:16206–16212. doi:10.1021/ja0650347
Zheng CH, Zhou YJ, Zhu J et al (2007) Bioorg Med Chem 15:6407–6417. doi:10.1016/j.bmc.2007.06.052
Pinto M, Perez JJ, Rubio-Martinez J (2004) J Comput Aided Mol Des 18:13–22. doi:10.1023/B:JCAM.0000022559.72848.1c
Mancinelli F, Caraglia M, Budillon A et al (2006) J Cell Biochem 99:305–318. doi:10.1002/jcb.20893
Fu XR, Apgar JR, Keating AE (2007) J Mol Biol 371:1099–1117. doi:10.1016/j.jmb.2007.04.069
Eyrisch S, Helms V (2007) J Med Chem 50:3457–3464. doi:10.1021/jm070095g
Manion MK, O’Neill JW, Giedt CD et al (2004) J Biol Chem 279:2159–2165. doi:10.1074/jbc.M306021200
Berendsen HJC, Vanderspoel D, Vandrunen R (1995) Comput Phys Commun 91:43–56. doi:10.1016/0010-4655(95)00042-E
Lindahl E, Hess B, van der Spoel D (2001) J Mol Model 7:306–317
Van der Spoel D, Lindahl E, Hess B et al (2005) J Comput Chem 26:1701–1718. doi:10.1002/jcc.20291
Jorgensen WL, Maxwell DS, TiradoRives J (1996) J. Am. Chem. Soc 118:11225–11236. doi:10.1021/ja9621760
Kaminski GA, Friesner RA, Tirado-Rives J et al (2001) J Phys Chem B 105:6474–6487. doi:10.1021/jp003919d
Jorgensen WL, Chandrasekhar J, Madura JD et al (1983) J Chem Phys 79:926–935. doi:10.1063/1.445869
Essmann U, Perera L, Berkowitz ML et al (1995) J Chem Phys 103:8577–8593. doi:10.1063/1.470117
Hoover WG (1985) Phys Rev A 31:1695–1697. doi:10.1103/PhysRevA.31.1695
Berendsen HJC, Postma JPM, Vangunsteren WF et al (1984) J Chem Phys 81:3684–3690. doi:10.1063/1.448118
Ghosh A, Rapp CS, Friesner RA (1998) J Phys Chem B 102:10983–10990. doi:10.1021/jp982533o
den Otter WK, Briels WJ (1998) J Chem Phys 109:4139–4146. doi:10.1063/1.477019
Ryckaert JP, Ciccotti G, Berendsen HJC (1977) J Comput Phys 23:327–341. doi:10.1016/0021-9991(77)90098-5
Lugovskoy AA, Degterev AI, Fahmy AF et al (2002) J Am Chem Soc 124:1234–1240. doi:10.1021/ja011239y
McGaughey GB, Gagne M, Rappe AK (1998) J Biol Chem 273:15458–15463. doi:10.1074/jbc.273.25.15458
Acknowledgments
This work was funded by a grant from Boston College to G.K.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Novak, W., Wang, H. & Krilov, G. Role of protein flexibility in the design of Bcl-XL targeting agents: insight from molecular dynamics. J Comput Aided Mol Des 23, 49–61 (2009). https://doi.org/10.1007/s10822-008-9237-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-008-9237-0