Abstract
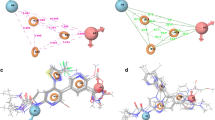
Tuberculosis (TB) is the primary cause of mortality among infectious diseases. Mycobacterium tuberculosis monophosphate kinase (TMPKmt) is essential to DNA replication. Thus, this enzyme represents a promising target for developing new drugs against TB. In the present study, the receptor-independent, RI, 4D-QSAR method has been used to develop QSAR models and corresponding 3D-pharmacophores for a set of 81 thymidine analogues, and two corresponding subsets, reported as inhibitors of TMPKmt. The resulting optimized models are not only statistically significant with r 2 ranging from 0.83 to 0.92 and q 2 from 0.78 to 0.88, but also are robustly predictive based on test set predictions. The most and the least potent inhibitors in their respective postulated active conformations, derived from each of the models, were docked in the active site of the TMPKmt crystal structure. There is a solid consistency between the 3D-pharmacophore sites defined by the QSAR models and interactions with binding site residues. Moreover, the QSAR models provide insights regarding a probable mechanism of action of the analogues.





Similar content being viewed by others
References
Dye C, Floyd K, Uplekar M (2008) World Health Organization Document, WHO/HTM/TB/2008.393
Aziz MA, Wright A, Laszlo A, De Muynck A, Portaels F, Van Deun A, Wells C, Nunn P, Blanc L, Raviglione M (2006) Lancet 368:2142–2154
Dye C (2006) Lancet 367:938–940
Andrade CH, Salum LB, Pasqualoto KFM, Ferreira EI, Andricopulo AD (2008) Lett Drug Des Discov 5:377–387
Andrade CH, Pasqualoto KFM, Zaim MH, Ferreira EI (2008) Braz J Pharm Sci 44:167–179
Haouz A, Vanheusden V, Munier-Lehmann H, Froeyen M, Herdewijn P, Van Calenbergh S, Delarue M (2003) J Biol Chem 278:4963–4971
Munier-Lehmann H, Chafotte A, Pochet S, Labesse G (2001) Protein Sci 10:1195–1205
Li de la Sierra I, Munier-Lehmann H, Gilles AM, Bârzu O, Delarue M (2001) J Mol Biol 311:87–100
Vanheusden V, Munier-Lehmann H, Froeyen M, Busson R, Rozenski J, Herdewijn P, Van Calenbergh S (2004) J Med Chem 47:6187–6194
Vanheusden V, Munier-Lehmann H, Pochet S, Herdewijn P, Van Calenbergh S (2002) Bioorg Med Chem Lett 12:2695–2698
Vanheusden V, Van Rompaey P, Munier-Lehmann H, Pochet S, Herdewijn P, Van Calenbergh S (2003) Bioorg Med Chem Lett 13:3045–3048
Van Daele I, Munier-Lehmann H, Froeyen M, Balzarini J, Van Calenbergh S (2007) J Med Chem 50:5281–5292
Gopalakrishnan B, Aparna V, Jeevan J, Ravi M, Desiraju GR (2005) J Chem Inf Model 45:1101–1108
Aparna V, Jeevan J, Ravi M, Desiraju GR, Gopalakrishnan B (2006) Bioorg Med Chem Lett 16:1014–1020
Andrade CH, Pasqualoto KFM, Ferreira EI, Hopfinger AJ (2009) J Chem Inf Model 49:1070–1078
Hopfinger AJ, Wang S, Tokarski JS, Jin B, Albuquerque M, Madhav PJ, Duraiswami C (1997) J Am Chem Soc 119:10509–10524
Blondin C, Serina L, Wiesmuller L, Gilles AM, Barzu (1994) Anal Biochem 220:219–222
HyperChem Program Release 7.05 for Windows (2005) Hybercube Inc. Gainesville, FL
Dewar MJSE, Zoebisch G, Healy EF, Stewart JJP (1985) J Am Chem Soc 107:3902–3909
4D-QSAR Package version 2.0 (1997) The Chem21 Group Inc. Lake Forest, IL
Pasqualoto KFM, Ferreira EI, Santos OAF, Hopfinger AJ (2004) J Med Chem 47:3755–3764
Romeiro NC, Albuquerque MG, Alencastro RB, Ravi M, Hopfinger AJ (2005) J Comput Aided Mol Des 19:385–400
Doherty DC (2001) MOLSIM Package version 3.2. The Chem21 Group Inc, Lake Forest, IL
Ghose AK, Pritchett A, Crippen GM (1988) J Comput Chem 9:80–90
Glen WG, Dunn WJ III, Scott DR (1989) Tetrahedron Comput Methodol 2:349–354
Rogers DG, Hopfinger AJ (1994) J Chem Inf Comput Sci 34:854–866
Dunn WJ III, Rogers D (1996) In: Devillers J (ed) Genetic algorithms in molecular modeling. Academic Press, London
Friedman JH (1991) Ann Stat 19:1–141
Discovery Studio Visualizer version 2.0 (2007) Accelrys Software Inc. San Diego, CA. http://accelrys.com/
DeLano WL (2004) The Pymol Molecular Graphics System version 1.0. Delano Scientific LLC: Palo Alto, CA. http://www.pymol.org/
Acknowledgments
The authors are grateful to the CAPES Foundation, a federal scientific agency of Brazil, for scholarship support. This work was also funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant 1 R21 GM075775. Information on Novel Preclinical Tools for Predictive ADME-Toxicology can be found at http://grants.nih.gov/grants/guide/rfa-files/RFA-RM-04-023.html. Links to nine initiatives are found here http://nihroadmap. nih.gov/initiatives.asp. Resources of the Laboratory of Molecular Modeling and Design at UNM and The Chem21 Group, Inc. were used in performing this work.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Andrade, C.H., Pasqualoto, K.F.M., Ferreira, E.I. et al. 3D-Pharmacophore mapping of thymidine-based inhibitors of TMPK as potential antituberculosis agents. J Comput Aided Mol Des 24, 157–172 (2010). https://doi.org/10.1007/s10822-010-9323-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-010-9323-y