Abstract
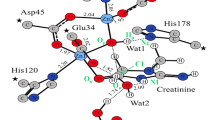
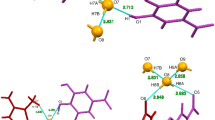
The reaction mechanism of creatinine-creatininase binding to form creatine as a final product has been investigated by using a combined ab initio quantum mechanical/molecular mechanical approach and classical molecular dynamics (MD) simulations. In MD simulations, an X-ray crystal structure of the creatininase/creatinine was modified for creatininase/creatinine complexes and the MD simulations were run for free creatininase and creatinine in water. MD results reveal that two X-ray water molecules can be retained in the active site as catalytic water. The binding free energy from Molecular Mechanics Poisson-Boltzmann Surface Area calculation predicted the strong binding of creatinine with Zn2+, Asp45 and Glu183. Two step mechanisms via Mn2+/Zn2+ (as in X-ray structure) and Zn2+/Zn2+ were proposed for water adding step and ring opening step with two catalytic waters. The pathway using synchronous transit methods with local density approximations with PWC functional for the fragment in the active region were obtained. Preferable pathway Zn2+/Zn2+ was observed due to lower activation energy in water adding step. The calculated energy in the second step for both systems were comparable with the barrier of 26.03 and 24.44 kcal/mol for Mn2+/Zn2+ and Zn2+/Zn2+, respectively.




Similar content being viewed by others
References
Spencer K (1986) Analytical reviews in clinical biochemistry: the estimation of creatinine. Ann Clin Biochem 23:1–25
Narayana S, Appleton HD (1986) Creatinine: a review. Clin Chem 26:1119–1126
Tietz NW (1986) Textbook of clinical chemistry. Saunders, Philadelphia, pp 1810–1857
Sena SF, Syed D, McComb RB (1988) Effect of high creatine content on the Kodak single-slide method for creatinine. Clin Chem 34:594–595
Pandey PC, Mishra AP (2004) Novel potentiometric sensing of creatinine. Sensor Actuator B Chem 99:230–235
Premanode B, Toumazou C (2007) A novel, low power biosensor for real time monitoring of creatinine and urea in peritoneal dialysis. Sensor Actuator B Chem 120:732–735
Yoshimoto T, Tanaka N, Kanada N, Inoue T, Nakajima Y, Haratake M, Nakamura KT, Xu Y, Ito K (2004) Crystal structures of creatininase reveal the substrate binding site and provide an insight into the catalytic mechanism. J Mol Biol 337:399–416
Tanaka N, Kusakabe Y, Ito K, Yoshimoto T, Nakamura KT (2002) Crystal structure of formaldehyde dehydrogenase from Pseudomonas putida: the structural origin of the tightly bound cofactor in nicotinoprotein dehydrogenases. J Mol Biol 324:519–533
Ito K, Kanada N, Inoue T, Furukawa K, Yamashita K, Tanaka N, Nakamura KT, Nishiya Y, Sogabe A, Yoshimoto T (2002) Preliminary crystallographic studies of the creatinine amidohydrolase from Pseudomonas putida. Acta Crystallogr D 58:2180–2181
Case DA, Cheatham Iii TE, Darden T, Gohlke H, Luo R, Merz KM Jr, Onufriev A, Simmerling C, Wang B, Woods RJ (2005) The Amber biomolecular simulation programs. J Comput Chem 26:1668–1688
Massova I, Kollman PA (2000) Combined molecular mechanical and continuum solvent approach (MM- PBSA/GBSA) to predict ligand binding, Perspect. Drug Discov Des 18:113–135
Wang JM, Hou T, Xu X (2006) Recent advances in free energy calculations with a combination of molecular mechanics and continuum models. Curr Comput Aided Drug Des 2:287–306
Bradbrook GM, Gleichmann T, Harrop SJ, Habash J, Raftery J, Kalb J, Yariv J, Hillier IH, Helliwell JR (1998) X-ray and molecular dynamics studies of concanavalin-A glucoside and mannoside complexes: Relating structure to thermodynamics of binding. J Chem Soc Faraday Trans 94:1603–1611
Merz KM Jr (1991) CO2 binding to human carbonic anhydrase II. J Am Chem Soc 113:406–411
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Zakrzewski VG, Montgomery JA, Stratmann RE, Burant JC, Dapprich S, Millam JM, Daniels AD, Kudin KN, Strain MC, Farkas O, Tomasi J, Barone V, Cossi M, Cammi R, Menucci B, Pomelli C, Adamo C, Clifford S, Ochterski J, Petersson GA, Ayala PY, Cui Q, Morokuma K, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Cioslowki J, Ortiz JV, Stefanov BB, Liu A, Liashenko A, Piskorz P, Komaromi I, Gomperts R, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Gonzalez M, Callacombe M, Gill PMW, Johnson BG, Chen W, Wong MW, Andres JL, Head-Gordon M, Replogle ES, Pople JA, Gaussian03 Program (1998)
Accelrys Software Inc (2007) Materials modeling and simulation software, release 4.3. Accelrys Software Inc, San Diego
Delley BJ (1990) DMOL is available commercially from BIOSYM technologies, San Diego, CA. Chem Phys 92:508
Delley BJ (2000) Dmol3 is available as part of materials studio. Chem Phys 113:7756
Perdew JP, Wang Y (1992) Accurate and simple analytic representation of the electron-gas correlation energy. Phys Rev B 45:13244–13249
Halgren TA, Lipscomb WN (1977) The synchronous-transit method for determining reaction pathways and locating molecular transition states. Chem Phys Lett 49:225–232
Acknowledgments
The authors would like to express grateful acknowledgement to the Thailand Research Fund (TRF), Computational Nanoscience Consortium (CNC), and National Nanotechnology Center (NANOTEC Thailand) for the access to Discovery Studio Version 1.7 program package. The authors also acknowledge a financial support from the Center for Innovation in Chemistry (PERCH-CIC), and Commission on Higher Education, Ministry of Education of Thailand.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lee, V.S., Kodchakorn, K., Jitonnom, J. et al. Influence of metal cofactors and water on the catalytic mechanism of creatininase-creatinine in aqueous solution from molecular dynamics simulation and quantum study. J Comput Aided Mol Des 24, 879–886 (2010). https://doi.org/10.1007/s10822-010-9380-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-010-9380-2