Abstract
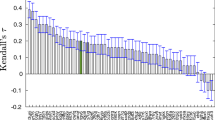
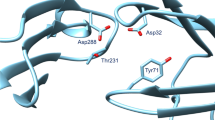
In this work we propose a protocol for estimating the effect of pH on the docking performance to BACE-1, which affords the charge state of the inhibitor as well as the protonation state of all ionisable residues in the protein at a given pH value. To the best of our knowledge, this is the first report of a protocol predicting the BACE-1 ligand docking poses not only at the neutral pH at which most crystallographic structures were obtained, but also at the optimal pH of the enzyme (in the acidic range), at which most of the BACE-1 binding affinity assays are performed. We have applied this protocol to a set of 23 fragment-like BACE-1 ligands that span four orders of magnitude in their binding affinities. The pK a values of the BACE-1 acidic residues deviate substantially from the estimates for model compounds in solution and display a ligand dependent variability, especially in the case of the catalytic Asp dyad residues. This outcome should have a strong bearing on the design of protocols for docking based BACE-1 screening campaigns. Finally, we were able to find an explanation for the poor docking success rate of some fragments based on the availability of anchoring points, a rationale that could help to improve hit rates in BACE-1 screening campaigns.







Similar content being viewed by others
References
Selkoe DJ (2011) Resolving controversies on the path to Alzheimer’s therapeutics. Nat Med 17:1060–1065
Villaverde MC, González-Louro L, Sussman F (2007) The search for drug leads targeted to the β-secretase: an example of the roles of computer assisted approaches in drug discovery. Curr Top Med Chem 7:980–990
Grüninger-Leitch F, Schlatter D, Küng E, Nelböck P, Döbeli H (2002) Substrate and inhibitor profile of BACE (β-secretase) and comparison with other mammalian aspartic proteases. J Biol Chem 277:4687–4693
Coburn CA, Stachel SJ, Li YM, Rush DM, Steele TG, Chen-Dodson E, Holloway MK, Xu M, Huang Q, Lai MT, DiMuzio J, Crouthamel MC, Shi XP, Sardana V, Chen Z, Munshi S, Kuo L, Makara GM, Annis DA, Tadikonda PK, Nash HM, Vacca JP, Wang T (2004) Identification of a small molecule nonpeptide active site β-secretase inhibitor that displays a nontraditional binding mode for aspartyl proteases. J Med Chem 47:6117–6119
Murray CW, Callaghan O, Chessari G, Cleasby A, Congreve M, Frederickson M, Hartshorn MJ, McMenamin R, Patel S, Wallis N (2007) Application of fragment screening by X-ray crystallography to β-secretase. J Med Chem 50:1116–1123
Congreve M, Aharony D, Albert J, Callaghan O, Campbell J, Carr RAE, Chessari G, Cowan S, Edwards PD, Frederickson M, McMenamin R, Murray CW, Patel S, Wallis N (2007) Application of fragment screening by X-ray crystallography to the discovery of aminopyridines as inhibitors of β-secretase. J Med Chem 50:1124–1132
Edwards PD, Albert JS, Sylvester M, Aharony D, Andisik D, Callaghan O, Campbell JB, Carr RA, Chessari G, Congreve M, Frederickson M, Folmer RHA, Geschwindner S, Koether G, Kolmodin K, Krumrine J, Mauger RC, Murray CW, Olsson LL, Patel S, Spear N, Tian G (2007) Application of fragment-based lead generation to the discovery of novel, cyclic amidine β-secretase inhibitors with nanomolar potency, cellular activity, and high ligand efficiency. J Med Chem 50:5912–5925
Yang W, Fucini RV, Fahr BT, Randal M, Lind KE, Lam MB, Lu W, Lu Y, Cary DR, Romanowski MJ, Colussi D, Pietrak B, Allison TJ, Munshi SK, Penny DM, Pham P, Sun J, Thomas AE, Wilkinson JM, Jacobs JW, McDowell RS, Ballinger MD (2009) Fragment-based discovery of nonpeptidic BACE-1 inhibitors using tethering. Biochemistry 48:4488–4499
Kuglstatter A, Stahl M, Peters JW, Huber W, Stihle M, Schlatter D, Benz J, Ruf A, Roth D, Enderle T, Hennig M (2008) Tyramine fragment binding to BACE-1. Bioorg Med Chem Lett 18:1304–1307
Wang YS, Strickland C, Voigt JH, Kennedy ME, Beyer BM, Senior MM, Smith EM, Nechuta TL, Madison VS, Czarniecki M, McKittrick BA, Stamford AW, Parker EM, Hunter JC, Greenlee WJ, Wyss DF (2010) Application of fragment-based NMR screening, X-ray crystallography, structure-based design, and focused chemical library design to identify novel μM leads for the development of nM BACE-1 (β-site APP cleaving enzyme 1) inhibitors. J Med Chem 53:942–950
Madden J, Dod JR, Godemann R, Kraemer J, Smith M, Biniszkiewicz M, Hallett DJ, Barker J, Dyekjaer JD, Hesterkamp T (2010) Fragment-based discovery and optimization of BACE1 inhibitors. Bioorg Med Chem Lett 20:5329–5333
Stachel SJ, Coburn CA, Rush D, Jones KLG, Zhu H, Rajapakse H, Graham SL, Simon A, Holloway MK, Allison TJ, Munshi SK, Espeseth AS, Zuck P, Colussi D, Wolfe A, Pietrak BL, Lai MT, Vacca JP (2009) Discovery of aminoheterocycles as a novel β-secretase inhibitor class: pH dependence on binding activity part 1. Bioorg Med Chem Lett 19:2977–2980
Huang D, Lüthi U, Kolb P, Cecchini M, Barberis A, Caflisch A (2006) In silico discovery of β-secretase inhibitors. J Am Chem Soc 128:5436–5443
Polgár T, Keserü GM (2005) Virtual screening for β-secretase (BACE1) inhibitors reveals the importance of protonation states at Asp32 and Asp228. J Med Chem 48:3749–3755
Polgár T, Magyar C, Simon I, Keserü GM (2007) Impact of ligand protonation on virtual screening against β-secretase (BACE1). J Chem Inf Model 47:2366–2373
Domínguez JL, Christopeit T, Villaverde MC, Gossas T, Otero JM, Nyström S, Baraznenok V, Lindström E, Danielson UH, Sussman F (2010) Effect of the protonation state of the titratable residues on the inhibitor affinity to BACE-1. Biochemistry 49:7255–7263
Sussman F, Otero JM, Villaverde MC, Castro M, Domínguez JL, González-Louro L, Estévez RJ, Estévez JC (2011) On a possible neutral charge state for the catalytic dyad in β-secretase when bound to hydroxyethylene transition state analogue inhibitors. J Med Chem 54:3081–3085
Kacker P, Masetti M, Mangold M, Bottegoni G, Cavalli A (2012) Combining dyad protonation and active site plasticity in BACE-1 structure-based drug design. J Chem Inf Model 52:1079–1085
Barman A, Prabhakar R (2012) Protonation states of the catalytic dyad of β-secretase (BACE1) in the presence of chemically diverse inhibitors: a molecular docking study. J Chem Inf Model 52:1275–1287
Discovery Studio, version 2.1, Accelrys Inc., San Diego, CA
Spassov VZ, Yan L (2008) A fast and accurate computational approach to protein ionization. Protein Sci 17:1955–1970
Im W, Lee MS, Brooks CL III (2003) Generalized born model with a simple smoothing function. J Comput Chem 24:1691–1702
SciFinder results calculated using ACD/Labs software V11.02. Advanced Chemistry Development, Inc., Toronto, ON
GOLD, version 5.1. Cambridge Crystallographic Data Centre, Cambridge
Jones G, Willett P, Glen RC (1995) Molecular recognition of receptor sites using a genetic algorithm with a description of desolvation. J Mol Biol 245:43–53
Jones G, Willett P, Glen RC, Leach AR, Taylor R (1997) Development and validation of a genetic algorithm for flexible docking. J Mol Biol 267:727–748
Baxter CA, Murray CW, Clark DE, Westhead DR, Eldridge MD (1998) Flexible docking using tabu search and an empirical estimate of binding affinity. Proteins 33:367–382
Eldridge MD, Murray CW, Auton TR, Paolini GV, Mee RP (1997) Empirical scoring functions: I. The development of a fast empirical scoring function to estimate the binding affinity of ligands in receptor complexes. J Comput Aided Mol Des 11:425–445
Verdonk ML, Cole JC, Hartshorn M, Murray CW, Taylor RD (2003) Improved protein-ligand docking using GOLD. Proteins 52:609–623
Korb O, Stützle T, Exner TE (2009) Empirical scoring functions for advanced protein-ligand docking with PLANTS. J Chem Inf Model 49:84–96
Hoffmann D, Kramer B, Washio T, Steinmetzer T, Rarey M, Lengauer T (1999) Two-stage method for protein-ligand docking. J Med Chem 42:4422–4433
Gleeson MP, Gleeson D (2009) QM/MM as a tool in fragment based drug discovery. A cross-docking, rescoring study of kinase inhibitors. J Chem Inf Model 49:1437–1448
Momany FA, Klimkowski VJ, Schäfer L (1990) On the use of conformationally dependent geometry trends from ab initio dipeptide studies to refine potentials for the empirical force field CHARMM. J Comput Chem 11:654–662
Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M (1983) CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem 4:187–217
Verdonk ML, Giangreco I, Hall RJ, Korb O, Mortenson PN, Murray CW (2011) Docking performance of fragments and drug like compounds. J Med Chem 54:5422–5431
Graves AP, Shivakumar DM, Boyce SE, Jacobson MP, Case DA, Shoichet BK (2008) Rescoring docking hit lists for model cavity sites: predictions and experimental testing. J Mol Biol 377:914–934
Brenk R, Vetter SW, Boyce SE, Goodin DB, Shoichet BK (2006) Probing molecular docking in a charged model binding site. J Mol Biol 357:1449–1470
Acknowledgments
This work was supported by financial aid from the Xunta de Galicia (Grant PGIDIT 10CSA209063PR) and Ministerio de Educación y Ciencia fellowship to J.L.D. The Supercomputing Center of Galicia (CESGA) provided computer time.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Domínguez, J.L., Villaverde, M.C. & Sussman, F. Effect of pH and ligand charge state on BACE-1 fragment docking performance. J Comput Aided Mol Des 27, 403–417 (2013). https://doi.org/10.1007/s10822-013-9653-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-013-9653-7