Abstract
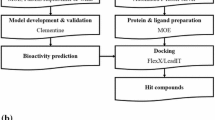
P-glycoprotein (P-gp) is an ATP-binding cassette multidrug transporter. The over expression of P-gp leads to the development of multidrug resistance (MDR), which is a major obstacle to effective treatment of cancer. Thus, designing effective P-gp inhibitors has an extremely important role in the overcoming MDR. In this paper, both ligand-based quantitative structure–activity relationship (QSAR) and receptor-based molecular docking are used to predict P-gp inhibitors. The results show that each method achieves good prediction performance. According to the results of tenfold cross-validation, an optimal linear SVM model with only three descriptors is established on 857 training samples, of which the overall accuracy (Acc), sensitivity, specificity, and Matthews correlation coefficient are 0.840, 0.873, 0.813, and 0.683, respectively. The SVM model is further validated by 418 test samples with the overall Acc of 0.868. Based on a homology model of human P-gp established, Surflex-dock is also performed to give binding free energy-based evaluations with the overall accuracies of 0.823 for the test set. Furthermore, a consensus evaluation is also performed by using these two methods. Both QSAR and molecular docking studies indicate that molecular volume, hydrophobicity and aromaticity are three dominant factors influencing the inhibitory activities.





Similar content being viewed by others
References
Nobili S, Landini I, Mazzei T, Mini E (2012) Overcoming tumor multidrug resistance using drugs able to evade P-glycoprotein or to exploit its expression. Med Res Rev 32(6):1220–1262
Wu C-P, Ohnuma S, Ambudkar SV (2011) Discovering natural product modulators to overcome multidrug resistance in cancer chemotherapy. Curr Pharm Biotechnol 12(4):609
Alexiou GA, Goussia A, Kyritsis AP, Tsiouris S, Ntoulia A, Malamou-Mitsi V, Voulgaris S, Fotopoulos AD (2011) Influence of glioma’s multidrug resistance phenotype on 99mTc-tetrofosmin uptake. Mol Imag Biol 13(2):348–351
Li W, Samra DA, Merzaban J, Khashab NM (2013) P-glycoprotein targeted nanoscale drug carriers. J Nanosci Nanotechnol 13(2):1399–1402
Gong J, Jaiswal R, Mathys J-M, Combes V, Grau G, Bebawy M (2012) Microparticles and their emerging role in cancer multidrug resistance. Cancer Treat Rev 38(3):226–234
Zhao L, Sun Y, Li X, Jin X, Xu Y, Guo Z, Liang R, Ding X, Chen T, Wang S (2011) Multidrug resistance strength of the novel multidrug resistance gene HA117: compared with MRP1. Med Oncol 28(4):1188–1195
Nobili S, Landini I, Mazzei T, Mini E (2011) Overcoming tumor multidrug resistance using drugs able to evade P-glycoprotein or to exploit its expression. Med Res Rev 32:1220–1262
Xiong X-B, Lavasanifar A (2011) Traceable multifunctional micellar nanocarriers for cancer-targeted co-delivery of MDR-1 siRNA and doxorubicin. ACS Nano 5(6):5202–5213
Wang X, Li J, Wang Y, Koenig L, Gjyrezi A, Giannakakou P, Shin EH, Tighiouart M, Chen Z, Nie S (2011) A folate receptor-targeting nanoparticle minimizes drug resistance in a human cancer model. ACS Nano 5(8):6184–6194
Shtil AA (2001) Signal transduction pathways and transcriptional mechanisms as targets for prevention of emergence of multidrug resistance in human cancer cells. Curr Drug Targets 2(1):57–77
Liu M, Hou T, Feng Z, Li Y (2012) The flexibility of P-glycoprotein for its poly-specific drug binding from molecular dynamics simulations. J Biomol Struct Dyn (ahead-of-print):1–18
Bikadi Z, Hazai I, Malik D, Jemnitz K, Veres Z, Hari P, Ni ZL, Loo TW, Clarke DM, Hazai E, Mao QC (2011) Predicting P-glycoprotein-mediated drug transport based on support vector machine and three-dimensional crystal structure of P-glycoprotein. PLoS ONE 6(10):e25815
Muller H, Pajeva IK, Globisch C, Wiese M (2008) Functional assay and structure-activity relationships of new third-generation P-glycoprotein inhibitors. Bioorg Med Chem 16(5):2448–2462
Pajeva IK, Globisch C, Wiese M (2009) Combined pharmacophore modeling, docking, and 3D QSAR studies of ABCB1 and ABCC1 transporter inhibitors. Chem Med Chem 4(11):1883–1896
Chen L, Li Y, Zhao Q, Peng H, Hou T (2011) ADME evaluation in drug discovery. 10. Predictions of P-glycoprotein inhibitors using recursive partitioning and naive Bayesian classification techniques. Mol Pharm 8(3):889–900
Sirisha K, Shekhar MC, Umasankar K, Mahendar P, Sadanandam A, Achaiah G, Reddy VM (2011) Molecular docking studies and in vitro screening of new dihydropyridine derivatives as human MRP1 inhibitors. Bioorg Med Chem 19(10):3249–3254
Klepsch F, Chiba P, Ecker GF (2011) Exhaustive sampling of docking poses reveals binding hypotheses for propafenone type inhibitors of P-glycoprotein. PLoS Comput Biol 7(5):e1002036
Becker JP, Depret G, Van Bambeke F, Tulkens PM, Prevost M (2009) Molecular models of human P-glycoprotein in two different catalytic states. BMC Struct Biol 9:3
Broccatelli F, Carosati E, Neri A, Frosini M, Goracci L, Oprea TI, Cruciani G (2011) A novel approach for predicting P-glycoprotein (ABCB1) inhibition using molecular interaction fields. J Med Chem 54(6):1740–1751
Sybyl 8.1; Tripos Inc.: St. Louis M, USA, 2008; Available online: http://www.tripos.com
Kalita MK, Nandal UK, Pattnaik A, Sivalingam A, Ramasamy G, Kumar M, Raghava GPS, Gupta D (2008) CyclinPred: a SVM-based method for predicting cyclin protein sequences. PLoS ONE 3(7):e2605
Pan XC, Mei H, Xie JA, Lu J, Wang Q, Zhang YL, Tan W (2012) Prediction of TAP binding affinity of peptide and selection specificity using VHSE descriptors. Chem J Chin Univ-Chin 33(11):2556–2562
Xie JA, Mei H, Lu J, Pan XC, Wang Q, Zhang YL (2012) Studies on the prediction of selective cleavage sites and cleavage profile of proteasome using VHSE amino acid descriptor. Acta Chim Sinica 70(3):318–324
Zhang YL, Mei H, Wang Q, Xie JA, Lv J, Pan XC, Tan W (2012) Peptide binding specificities of HLA-B*5701 and B*5801. Sci China-Life Sci 55(9):818–825
Noble WS (2006) What is a support vector machine? Nat Biotechnol 24(12):1565–1567
Chang C–C, Lin C-J (2011) LIBSVM: a library for support vector machines. ACM Trans Intell Syst Technol (TIST) 2(3):27
Šali A, Webb B, Madhusudhan M, Shen M, Marti-Renom M, Eswar N, Alber F, Topf M, Oliva B, Fiser A (2008) MODELLER. A program for protein structure modeling. Version 9v4, University of California, San Francisco, CA
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26(2):283–291
Jain AN (2003) Surflex: fully automatic flexible molecular docking using a molecular similarity-based search engine. J Med Chem 46(4):499–511
Wu B, Sun J, Cheng SP, Gu JD, Li AM, Zhang XX (2011) Comparative analysis of binding affinities between styrene and mammalian CYP2E1 by bioinformatics approaches. Ecotoxicology 20(5):1041–1046
Kellenberger E, Rodrigo J, Muller P, Rognan D (2004) Comparative evaluation of eight docking tools for docking and virtual screening accuracy. Proteins 57(2):225–242
Kruszewski J, Krygowski TM (1972) Definition of aromaticity basing on the harmonic oscillator model [J]. Tetrahedron Lett 13(36):3839–3842
Poongavanam V, Haider N, Ecker GF (2012) Fingerprint-based in silico models for the prediction of P-glycoprotein substrates and inhibitors. Bioorg Med Chem 20:5388–5395
Jensen AR (1999) The g factor: the science of mental ability. Psicothema 11(2):445–446
C-l Chen, Chin JE, Ueda K, Clark DP, Pastan I, Gottesman M, Roninson I (1986) Internal Duplication and Homology with Bacterial Transport Proteins in the mdr1 (P-Glycoprotein). Cell 47:381–389
Hrycyna CA, Airan LE, Germann UA, Ambudkar SV, Pastan I, Gottesman MM (1998) Structural flexibility of the linker region of human P-glycoprotein permits ATP hydrolysis and drug transport. Biochemistry 37(39):13660–13673
Singh DV, Godbole MM, Misra K (2013) A plausible explanation for enhanced bioavailability of P-gp substrates in presence of piperine: simulation for next generation of P-gp inhibitors. J Mol Model 19(1):227–238
Kim IW, Peng XH, Sauna ZE, FitzGerald PC, Xia D, Muller M, Nandigama K, Ambudkar SV (2006) The conserved tyrosine residues 401 and 1044 in ATP sites of human P-glycoprotein are critical for ATP binding and hydrolysis: evidence for a conserved subdomain, the A-loop in the ATP-binding cassette. Biochemistry (Mosc) 45(24):7605–7616
Perkins NJ, Schisterman EF (2006) The inconsistency of “optimal” cutpoints obtained using two criteria based on the receiver operating characteristic curve. Am J Epidemiol 163(7):670–675
Tarcsay A, Keseru GM (2011) Homology modeling and binding site assessment of the human P-glycoprotein. Future Med Chem 3(3):297–307
Aller SG, Yu J, Ward A, Weng Y, Chittaboina S, Zhuo R, Harrell PM, Trinh YT, Zhang Q, Urbatsch IL, Chang G (2009) Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science 323(5922):1718–1722
Acknowledgments
This research was supported by the National Natural Science Foundation of China (No 61073135) and the “111” project of “Introducing Talents of Discipline to Universities”.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
10822_2013_9697_MOESM1_ESM.docx
The list of 87, 67, 43, 23, and 3 descriptors as well as the corresponding weights are provided in Table S1–S5. (DOCX 23 kb)
Rights and permissions
About this article
Cite this article
Tan, W., Mei, H., Chao, L. et al. Combined QSAR and molecule docking studies on predicting P-glycoprotein inhibitors. J Comput Aided Mol Des 27, 1067–1073 (2013). https://doi.org/10.1007/s10822-013-9697-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-013-9697-8