Abstract
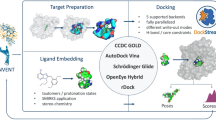
The SAMPL challenges provide an ideal opportunity for unbiased evaluation and comparison of different approaches used in computational drug design. During the fourth round of this SAMPL challenge, we participated in the virtual screening and binding pose prediction on inhibitors targeting the HIV-1 integrase enzyme. For virtual screening, we used well known and widely used in silico methods combined with personal in cerebro insights and experience. Regular docking only performed slightly better than random selection, but the performance was significantly improved upon incorporation of additional filters based on pharmacophore queries and electrostatic similarities. The best performance was achieved when logical selection was added. For the pose prediction, we utilized a similar consensus approach that amalgamated the results of the Glide-XP docking with structural knowledge and rescoring. The pose prediction results revealed that docking displayed reasonable performance in predicting the binding poses. However, prediction performance can be improved utilizing scientific experience and rescoring approaches. In both the virtual screening and pose prediction challenges, the top performance was achieved by our approaches. Here we describe the methods and strategies used in our approaches and discuss the rationale of their performances.








Similar content being viewed by others
References
Heikamp K, Bajorath J (2013) The future of virtual compound screening. Chem Biol Drug Des 81(1):33–40
Lill M (2013) Virtual screening in drug design. Methods Mol Biol 993:1–12
Geballe MT, Skillman AG, Nicholls A, Guthrie JP, Taylor PJ (2010) The SAMPL2 blind prediction challenge: introduction and overview. J Comput Aided Mol Des 24(4):259–279
Guthrie JP (2009) A blind challenge for computational solvation free energies: introduction and overview. J Phys Chem B 113(14):4501–4507
Nicholls A, Mobley DL, Guthrie JP, Chodera JD, Bayly CI, Cooper MD, Pande VS (2008) Predicting small-molecule solvation free energies: an informal blind test for computational chemistry. J Med Chem 51(4):769–779
Skillman AG (2012) SAMPL3: blinded prediction of host-guest binding affinities, hydration free energies, and trypsin inhibitors. J Comput Aided Mol Des 26(5):473–474
Voet AR, Maeyer MD, Debyser Z, Christ F (2009) In search of second-generation HIV integrase inhibitors: targeting integration beyond strand transfer. Future Med Chem 1(7):1259–1274
Peat TS, Rhodes DI, Vandegraaff N, Le G, Smith JA, Clark LJ, Jones ED, Coates JA, Thienthong N, Newman J, Dolezal O, Mulder R, Ryan JH, Savage GP, Francis CL, Deadman JJ (2012) Small molecule inhibitors of the LEDGF site of human immunodeficiency virus integrase identified by fragment screening and structure based design. PLoS ONE 7(7):e40147
Busschots K, Voet A, De Maeyer M, Rain JC, Emiliani S, Benarous R, Desender L, Debyser Z, Christ F (2007) Identification of the LEDGF/p75 binding site in HIV-1 integrase. J Mol Biol 365(5):1480–1492
Hombrouck A, De Rijck J, Hendrix J, Vandekerckhove L, Voet A, De Maeyer M, Witvrouw M, Engelborghs Y, Christ F, Gijsbers R, Debyser Z (2007) Virus evolution reveals an exclusive role for LEDGF/p75 in chromosomal tethering of HIV. PLoS Pathog 3(3):e47
Cavalluzzo C, Christ F, Voet A, Sharma A, Singh BK, Zhang KY, Lescrinier E, De Maeyer M, Debyser Z, Van der Eycken E (2013) Identification of small peptides inhibiting the integrase-LEDGF/p75 interaction through targeting the cellular co-factor. J Pept Sci 19(10):651–658
Cavalluzzo C, Voet A, Christ F, Singh BK, Sharma A, Debyser Z, Maeyer MD, Van der Eycken E (2012) De novo design of small molecule inhibitors targeting the LEDGF/p75-HIV integrase interaction. RSC Adv 2(3):974
Christ F, Voet A, Marchand A, Nicolet S, Desimmie BA, Marchand D, Bardiot D, Van der Veken NJ, Van Remoortel B, Strelkov SV, De Maeyer M, Chaltin P, Debyser Z (2010) Rational design of small-molecule inhibitors of the LEDGF/p75-integrase interaction and HIV replication. Nat Chem Biol 6(6):442–448
Kumar A, Voet A, Zhang KY (2012) Fragment based drug design: from experimental to computational approaches. Curr Med Chem 19(30):5128–5147
Rhodes DI, Peat TS, Vandegraaff N, Jeevarajah D, Le G, Jones ED, Smith JA, Coates JA, Winfield LJ, Thienthong N, Newman J, Lucent D, Ryan JH, Savage GP, Francis CL, Deadman JJ (2011) Structural basis for a new mechanism of inhibition of HIV-1 integrase identified by fragment screening and structure-based design. Antivir Chem Chemother 21(4):155–168
Peat TS, Dolezal O, Newman J, Mobley D, Deadman JJ (2014) Interrogating HIV integrase for compounds that bind—a SAMPL challenge. J Comput Aided Mol Des (this issue)
Cherepanov P, Ambrosio AL, Rahman S, Ellenberger T, Engelman A (2005) Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. Proc Natl Acad Sci USA 102(48):17308–17313
Wolber G, Langer T (2005) LigandScout: 3-D pharmacophores derived from protein–bound ligands and their use as virtual screening filters. J Chem Inf Model 45(1):160–169
OMEGA, version 2.4.6 OpenEye Scientific Software, Inc., Santa Fe, NM, USA. www.eyesopen.com, 2012
Hawkins PCD, Skillman AG, Warren GL, Ellingson BA, Stahl MT (2010) Conformer generation with OMEGA: algorithm and validation using high quality structures from the protein databank and cambridge structural database. J Chem Inf Model 50(4):572–584
Hawkins PC, Nicholls A (2012) Conformer generation with OMEGA: learning from the data set and the analysis of failures. J Chem Inf Model 52(11):2919–2936
Jones G, Willett P, Glen RC, Leach AR, Taylor R (1997) Development and validation of a genetic algorithm for flexible docking. J Mol Biol 267(3):727–748
Gehlhaar DK, Verkhivker GM, Rejto PA, Sherman CJ, Fogel DB, Fogel LJ, Freer ST (1995) Molecular recognition of the inhibitor AG-1343 by HIV-1 protease: conformationally flexible docking by evolutionary programming. Chem Biol 2(5):317–324
Molecular Operating Environment (MOE), version 2011.10; Chemical Computing Group Inc.: Montreal, Quebec, Canada, 2010
Voet A, Berenger F, Zhang KYJ (2013) Electrostatic similarities between protein and small molecule ligands facilitate the design of protein–protein interaction inhibitors. PLoS ONE 8(10):e75762
Glide, version 5.7, Schrödinger, LLC, New York, NY, 2011
Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS (2004) Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem 47(7):1739–1749
Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT (2006) Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein–ligand complexes. J Med Chem 49(21):6177–6196
Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, Banks JL (2004) Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J Med Chem 47(7):1750–1759
Maestro, version 9.2, Schrödinger, LLC, New York, NY, 2011
LigPrep, version 2.5, Schrödinger, LLC, New York, NY, 2011
Jorgensen WL, Maxwell DS, Tirado-Rives J (1996) Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc 118(45):11225–11236
Prime, version 3.0, Schrödinger, LLC, New York, NY, 2011
Bashford D, Case DA (2000) Generalized Born models of macromolecular solvation effects. Annu Rev Phys Chem 51(1):129–152
Tsui V, Case DA (2000) Theory and applications of the generalized born solvation model in macromolecular simulations. Biopolymers 56(4):275–291
Velec HFG, Gohlke H, Klebe G (2005) DrugScoreCSD-knowledge-based scoring function derived from small molecule crystal data with superior recognition rate of near-native ligand poses and better affinity prediction. J Med Chem 48(20):6296–6303
The PyMOL Molecular Graphics System, version 1.5.0.4 Schrödinger, LLC
Williams T, Kelley C (2012) Gnuplot 4.6: an interactive plotting program
Fry DC (2012) Small-molecule inhibitors of protein–protein interactions: how to mimic a protein partner. Curr Pharm Des 18(30):4679–4684
Voet A, Banwell EF, Sahu KK, Heddle JG, Zhang KY (2013) Protein interface pharmacophore mapping tools for small molecule protein: protein interaction inhibitor discovery. Curr Top Med Chem 13(9):989–1001
Voet A, Zhang KY (2012) Pharmacophore modelling as a virtual screening tool for the discovery of small molecule protein–protein interaction inhibitors. Curr Pharm Des 18(30):4586–4598
Mobley D, Liu S, Lim N, Deng N, Branson K, Perryman A, Forli S, Levy R, Gallicchio E, Olson A (2014) Blind prediction of HIV integrase binding from the SAMPL4 challenge. J Comput Aided Mol Des (this issue)
Voet A, Callewaert L, Ulens T, Vanderkelen L, Vanherreweghe JM, Michiels CW, De Maeyer M (2011) Structure based discovery of small molecule suppressors targeting bacterial lysozyme inhibitors. Biochem Biophys Res Commun 405(4):527–532
Voet A, Helsen C, Zhang KYJ, Claessens F (2013) The discovery of novel human androgen receptor antagonist chemotypes using a combined pharmacophore screening procedure. Chem Med Chem 8(4):644–651
Perryman AL, Santiago DN, Forli S, Olson AJ (2013) Virtual Screening with AutoDock Vina and the Common Pharmacophore Engine of a low diversity library of fragments and hits against the three allosteric sites of HIV integrase: participation in the SAMPL4 protein–ligand binding challenge. J Comput Aided Mol Des. doi:10.1007/s1082201497093
Coleman RG, Sterling T, Weiss DR (2013) SAMPL4 & DOCK3.7: Lessons for automated docking procedures. J Comput Aided Mol Des (in press)
Acknowledgments
We thank RIKEN Integrated Cluster of Clusters (RICC) at RIKEN for the supercomputing resources used for this study. We are grateful to Dr. David Mobley and SAMPL4 participants for sharing the SAMPL4 challenge analyses and prediction data prior to publication. We acknowledge the Initiative Research Unit program from RIKEN, Japan for funding. AV acknowledges the JSPS for a postdoctoral fellowship. Prof. Jeremy Tame is acknowledged for proofreading the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Arnout R. D. Voet and Ashutosh Kumar have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Voet, A.R.D., Kumar, A., Berenger, F. et al. Combining in silico and in cerebro approaches for virtual screening and pose prediction in SAMPL4. J Comput Aided Mol Des 28, 363–373 (2014). https://doi.org/10.1007/s10822-013-9702-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-013-9702-2