Abstract
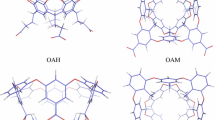
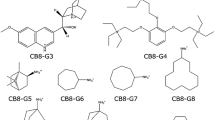
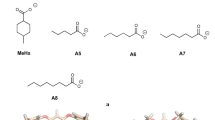
We have estimated free energies for the binding of nine cyclic carboxylate guest molecules to the octa-acid host in the SAMPL4 blind-test challenge with four different approaches. First, we used standard free-energy perturbation calculations of relative binding affinities, performed at the molecular-mechanics (MM) level with TIP3P waters, the GAFF force field, and two different sets of charges for the host and the guest, obtained either with the restrained electrostatic potential or AM1-BCC methods. Both charge sets give good and nearly identical results, with a mean absolute deviation (MAD) of 4 kJ/mol and a correlation coefficient (R 2) of 0.8 compared to experimental results. Second, we tried to improve these predictions with 28,800 density-functional theory (DFT) calculations for selected snapshots and the non-Boltzmann Bennett acceptance-ratio method, but this led to much worse results, probably because of a too large difference between the MM and DFT potential-energy functions. Third, we tried to calculate absolute affinities using minimised DFT structures. This gave intermediate-quality results with MADs of 5–9 kJ/mol and R 2 = 0.6–0.8, depending on how the structures were obtained. Finally, we tried to improve these results using local coupled-cluster calculations with single and double excitations, and non-iterative perturbative treatment of triple excitations (LCCSD(T0)), employing the polarisable multipole interactions with supermolecular pairs approach. Unfortunately, this only degraded the predictions, probably because of a mismatch between the solvation energies obtained at the DFT and LCCSD(T0) levels.










Similar content being viewed by others
References
Gohlke H, Klebe G (2002) Angew Chem Int Ed 41:2644
Jorgensen WL (2009) Acc Chem Res 42:724
Zhou H-X, Gilson MK (2009) Chem Rev 109:4092
Michel J, Essex JW (2010) J Comput Aided Mol Des 24:639
Christ CD, Mark AE, van Gunsteren WF (2010) J Comput Chem 31:1569
Wereszczynski J, McCammon JA (2012) Quart Rev Biophys 45:1
Halgren TA, Damm W (2001) Curr Opin Struct Biol 11:236
Söderhjelm P, Ryde U (2009) J Phys Chem A 113:617
Cavalli A, Carloni P, Recanatini M (2006) Chem Rev 106:3497
Werner H.-J, Knowles P. J, Knizia G, Manby F. R, Schütz M et al (2012) MOLPRO,, version 2012.1, a package of ab initio programs. see http://www.molpro.net
Raha K, Peters MB, Wang B, Yu N, Wollacott AM, Westerhoff LM, Merz KM (2007) Drug Discov Today 12:725
Söderhjelm P, Kongsted J, Genheden S, Ryde U (2010) Interdiscip Sci Comput Life Sci 2:21–37
Söderhjelm P, Genheden S, Ryde U (2012) Protein–ligand interactions. In: Gohlke H (ed) Methods and principles in medicinal chemistry, vol 53. Wiley-VCH, Weinheim, pp 121–143
Antony J, Grimme S (2012) J Comput Chem 33:1730
Muddana HS, Varnado CD, Bielawski CW, Urbach AR, Isaacs L, Geballe MT, Gilson MK (2012) J Comput Aided Mol Des 26:475
Muddana HS, Fenley AT, Mobley DL, Gilson MK (2014) Blind prediction of the host–guest binding affinities from the SAMPL4 challenge. J Comput-Aided Mol Design (in press)
Gibb CLD, Gibb BCJ (2004) Am Chem Soc 126:11408
Sun H, Gibb CLD, Gibb BC (2008) Supramol Chem 20:141
Gibb CLD, Gibb BC (2009) Tetrahedron 65:7240
Grimme S, Antony J, Ehrlich S, Krieg H (2010) J Chem Phys 132:154104
Grimme S (2012) Chem Eur J 18:9955
Hampel C, Werner H-J (1996) J Chem Phys 104:6286
Andrejić M, Mata RA, Ryde U, Söderhjelm P (2014) Chem Phys Chem (submitted)
Wang JM, Wolf RM, Caldwell KW, Kollman PA, Case DA (2004) J Comput Chem 25:1157–1174
Jorgensen WL, Chandrasekhar J, Madura JD, Impley RW, Klein ML (1983) J Chem Phys 79:926–935
Case DA, Darden TA, Cheatham TE III, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Roberts BP, Wang B, Hayik S, Roitberg A, Seabra G, Kolossvai I, Wong KF, Paesani F, Vanicek J, Liu J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Cai Q, Ye Q, Wang J, Hsieh M-J, Cui G, Roe DR, Mathews DH, Seetin MG, Sagui C, Babin V, Luchko T, Gusarov S, Kovalenko A, Kollman PA (2010) AMBER 11. University of California, San Francisco
Jakalian A, Bush BL, Jack DB, Bayly CI (2000) J Comput Chem 21:132–146
Jakalian A, Jack DB, Bayly CI (2002) J Comput Chem 23:1623–1641
Bayly CI, Cieplak P, Cornell WD, Kollman PA (1993) J Phys Chem 97:10269–10280
Besler BH, Merz KM, Kollman PA (1990) J Comput Chem 11:431–439
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Montgomery JA Jr, Peralta JE, Ogliaro F, Bearpark M, Heyd JJ, Brothers E, Kudin KN, Staroverov VN, Kobayashi R, Normand J, Raghavachari K, Rendell A, Burant JC, Iyengar SS, Tomasi J, Cossi M, Rega N, Millam JM, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Martin RL, Morokuma K, Zakrzewski VG, Voth GA, Salvador P, Dannenberg JJ, Dapprich S, Daniels AD, Farkas Ö, Foresman JB, Ortiz JV, Cioslowski J, Fox DJ (2009) Gaussian 09, revision A02. Gaussian Inc, Wallingford CT
Ryckaert JP, Ciccotti G, Berendsen HJC (1977) J Comput Phys 23:327–341
Berendsen HJC, Postma JPM, Van Gunsteren WF, Dinola A, Haak JR (1984) J Chem Phys 81:3684–3690
Darden T, York D, Pedersen L (1993) J Chem Phys 98:10089–10092
Genheden S, Ryde U (2011) J Comput Chem 32:187
Wu X, Brooks BR (2003) Chem Phys Lett 381:512–518
Tembe BL, McCammon JA (1984) Comp Chem 8:281–283
Bennett CH (1976) J Comput Phys 22:245–268
Shirts MR, Pande VS (2005) J Chem Phys 122:144107
Shirts MR, Chodera JD (2008) J Chem Phys 129:124105
Kirkwood JG (1935) J Chem Phys 3:300–313
Zwanzig RWJ (1954) Chem Phys 22:1420–1426
Steinbrecher T, Mobley DL, Case DA (2007) J Chem Phys 127:214108
Ahlrichs R, Bär M, Häser M, Horn H, Kölmel C (1989) Chem Phys Lett 162:165
Treutler O, Ahlrichs RJ (1995) Chem Phys 102:346
Tao J, Perdew JP, Staroverov VN, Scuseria GE (2003) Phys Rev Lett 91:146401
Becke AD (1988) Phys Rev A 38:3098–3100
Perdew JP (1986) Phys Rev B 33:8822–8824
Schäfer A, Huber C, Ahlrichs R (1994) J Chem Phys 100:5829
Weigend F, Ahlrichs R (2005) Phys Chem Chem Phys 7:3297–3305
Weigend F, Furche F, Ahlrichs R (2003) J Chem Phys 119:12753
Eichkorn K, Treutler O, Öhm H, Häser M, Ahlrichs R (1995) Chem Phys Lett 240:283–290
Eichkorn K, Weigend F, Treutler O, Ahlrichs R (1997) Theor Chem Acc 97:119–126
Sierka M, Hogekamp A, Ahlrichs R (2003) J Chem Phys 118:9136
Grimme S, Ehrlich S, Goerigk L (2011) J Comput Chem 32:1456–1465
http://www.thch.uni-bonn.de/tc/index.php?section=downloads&subsection=getd3
Klamt A, Schüürmann J (1993) J Chem Soc Perkin Trans 2:799–805
Schäfer A, Klamt A, Sattel D, Lohrenz JCW, Eckert F (2000) Phys Chem Chem Phys 2:2187–2193
Klamt A, Jonas V, Bürger T, Lohrenz JCW (1998) J Phys Chem 102:5074–5085
Klamt A (1995) J Phys Chem 99:2224
Eckert F, Klamt A (2002) AIChE J 48:369
Eckert F, Klamt A (2010) COSMOtherm, version C30, release 1301. COSMOlogic GmbH & Co KG, Leverkusen
Jensen F (1999) Introduction to computational chemistry. Wiley, Chichester, pp 298–303
Kaukonen M, Söderhjelm P, Heimdal J, Ryde U (2008) J Chem Theory Comput 4:985
Söderhjelm P, Husberg C, Strambi A, Olivucci M, Ryde U (2009) J Chem Theory Comput 5:649
Hu L, Eliasson J, Heimdal J, Ryde U (2009) J Phys Chem A 113:11793
Genheden S, Ryde U (2012) J Chem Theory Comput 8:1449
Wesolowski T, Warshel A (1994) J Phys Chem 98:5183–5187
Olsson MH, Hong G, Warshel A (2003) J Am Chem Soc 125:5025–5039
Wood RH, Yezdimer EM, Sakane S, Barriocanal JA, Doren DJJ (1999) Chem Phys 110:1329
Rod TH, Ryde U (2005) Phys Rev Lett 94:138302
Plotnikov NV, Kamerlin SCL, Warshel A (2011) J Phys Chem B 115:7950–7962
Woods CJ, Manby FR, Mulholland AJ (2008) J Chem Phys 128:014109
Beierlein FR, Michel J, Essex JW (2011) J Phys Chem B 115:4911–4926
König G, Boresch S (2011) J Comput Chem 32:1082
Dunning TH (1989) J Chem Phys 90:1007
Woon DE, Dunning TH (1993) J Chem Phys 98:1358
Polly R, Werner H-J, Manby FR, Knowles PJ (2004) Mol Phys 102:2311
Werner H-J, Manby FR, Knowles PJ (2003) J Chem Phys 118:8149
Weigend F (2002) Phys Chem Chem Phys 4:4285
Weigend F, Köhn A, Hättig C (2002) J Chem Phys 116:3175
Pipek J, Mezey PG (1989) J Chem Phys 90:4916–4926
Mata RA, Werner H-J (2007) Mol Phys 105:2753–2761
Dieterich JM, Werner H-J, Mata RA, Metz S, Thiel W (2010) J Chem Phys 132:035101
Helgaker T, Klopper W, Koch H, Noga J (1997) J Chem Phys 106:9639
Genheden S, Ryde U (2010) J Comput Chem 31:837–846
Pearlman DA, Charifson PS (2001) J Med Chem 44:3417
Mikulskis P, Genheden S, Rydberg P, Sandberg L, Olsen L, Ryde U (2012) J Comput-Aided Mol Design 26:527–554
Gibb CLD, Gibb B (2013) J Comput-Aided Mol Design. doi:10.1007/s10822-013-9690-2
Heimdal J, Ryde U (2012) Phys Chem Chem Phys 14:12592–12604
Genheden S, Nilsson I, Ryde U (2011) J Chem Inf Model 51:947–958
Grimme S (2006) J Comput Chem 27:1787–1799
Ryde U, Mata RA, Grimme S (2011) Dalton Trans 40:11176
Sure R, Antony J, Grimme S (2014) J Phys Chem B (submitted)
Sure R, Grimme S (2013) J Comput Chem 34:1672–1685
Acknowledgments
This investigation has been supported by Grants from the Swedish research council (project 2010-5025). The computations were performed on computer resources provided by the Swedish National Infrastructure for Computing (SNIC) at Lunarc at Lund University and HPC2N at Umeå University. The collaboration between the Universities of Lund and Göttingen has been carried out within the framework of the International Research Training Group 1422 Metal Sites in Biomolecules—Structures, Regulation, Mechanisms and M. A. is supported through a Ph.D. scholarship in this International Research Training Group. D. C. thanks FEBS for a short-term fellowship. We are grateful to Prof. Stefan Grimme for providing us with the thermo program.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mikulskis, P., Cioloboc, D., Andrejić, M. et al. Free-energy perturbation and quantum mechanical study of SAMPL4 octa-acid host–guest binding energies. J Comput Aided Mol Des 28, 375–400 (2014). https://doi.org/10.1007/s10822-014-9739-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-014-9739-x
Keywords
Profiles
- Samuel Genheden View author profile