Abstract
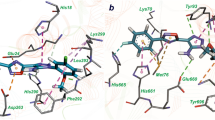
Mycobacterium tuberculosis infection remains a major cause of global morbidity and mortality due to the increase of antibiotics resistance. Dual/multi-target drug discovery is a promising approach to overcome bacterial resistance. In this study, we built ligand-based pharmacophore models and performed pharmacophore screening in order to identify hit compounds targeting simultaneously two enzymes—M. tuberculosis leucyl-tRNA synthetase (LeuRS) and methionyl-tRNA synthetase (MetRS). In vitro aminoacylation assay revealed five compounds from different chemical classes inhibiting both enzymes. Among them the most active compound—3-(3-chloro-4-methoxy-phenyl)-5-[3-(4-fluoro-phenyl)-[1,2,4]oxadiazol-5-yl]-3H-[1,2,3]triazol-4-ylamine (1) inhibits mycobacterial LeuRS and MetRS with IC50 values of 13 µM and 13.8 µM, respectively. Molecular modeling study indicated that compound 1 has similar binding mode with the active sites of both aminoacyl-tRNA synthetases and can be valuable compound for further chemical optimization in order to find promising antituberculosis agents.



Similar content being viewed by others
References
Francklyn CS, Mullen P (2019) Progress and challenges in aminoacyl-tRNA synthetase-based therapeutics. J Biol Chem 294:5365–5385
Kwon NH, Fox PL, Kim S (2019) Aminoacyl-tRNA synthetases as therapeutic targets. Nat Rev Drug Discov. https://doi.org/10.1038/s41573-019-0026-3
Ho JM, Bakkalbasi E, Söll D, Miller CA (2018) Drugging tRNA aminoacylation. RNA Biol 15:667–677
Rajendran V, Kalita P, Shukla H, Kumar A, Tripathi T (2018) Aminoacyl-tRNA synthetases: structure, function and drug discovery. Int J Biol Macromol 111:400–414
Randall CP, Rasina D, Jirgensons A, O’Neill AJ (2016) Targeting multiple aminoacyl-tRNA synthetases overcomes the resistance liabilities associated with antibacterial inhibitors acting on a single such enzyme. Antimicrob Agents Chemother 60:6359–6361
Li K, Shurig-Briccio LA, Feng X, Upadhyay A, Pujari V, Lechartier B, Fontes FL, Yang H, Rao G, Zhu W, Gulati A, No JH, Cintra G, Bogue S, Liu YL, Molohon K, Orlean P, Mitchell DA, Freitas-Junior L, Ren F, Sun H, Jiang T, Li Y, Guo RT, Cole ST, Gennis RB, Crick DC, Oldfield E (2014) Multitarget discovery for tuberculosis and other infectious diseases. J Med Chem 57:3126–3139
Gudzera OI, Golub AG, Bdzhola VG, Volynets GP, Lukashov SS, Kovalenko OP, Kriklivyi IA, Yaremchuk AD, Starosyla SA, Yarmoluk SM, Tukalo MA (2016) Discovery of potent anti-tuberculosis agents targeting leucyl-tRNA synthetase. Bioorg Med Chem 24:1023–1031
Li X, Hernandez V, Rock FL, Choi W, Mak YSL, Mohan M, Mao W, Zhou Y, Easom EE, Plattner JJ, Zou W, Pérez-Herrán E, Giordano I, Mendoza-Losana A, Alemparte C, Rullas J, Angulo-Barturen I, Crouch S, Ortega F, Barros D, Alley MRK (2017) Discovery of a potent and specific M. tuberculosis leucyl-tRNA synthetase inhibitor: (S)-3-(aminomethyl)-4-chloro-7-(2-hydroxyethoxy)benzo[c][1,2]oxaborol-1(3H)-ol (GSK656). J Med Chem 60:8011–8026
Faghih O, Zhang Z, Ranade RM, Gillespie JR, Creason SA, Huang W, Shibata S, Barros-Álvarez X, Verlinde CLMJ, Hol WGJ, Fan E, Buckner FS (2017) Development of methionyl-tRNA synthetase inhibitors as antibiotics for gram-positive bacterial infections. Antimicrob Agents Chemother. https://doi.org/10.1128/AAC.00999-17
Torrie LS, Brand S, Robinson DA, Ko EJ, Stojanovski L, Simeons FRC, Wyllie S, Thomas J, Ellis L, Osuna-Cabello M, Epemolu O, Nühs A, Riley J, MacLean L, Manthri S, Read KD, Gilbert IH, Fairlamb AH, De Rycker M (2017) Chemical validation of methionyl-tRNA synthetase as a druggable target in Leishmania donovani. ACS Infect Dis 3:718–727
Nayak SU, Griffiss JM, Blumer J, O’Riordan MA, Gray W, McKenzie R, Jurao RA, An AT, Le M, Bell SJ, Ochsner UA, Jarvis TC, Janjic N, Zenilman JM (2017) Safety, tolerability, systemic exposure, and metabolism of CRS3123, a methionyl-tRNA synthetase inhibitor developed for treatment of Clostridium difficile in phase I study. Antimicrob Agents Chemother. https://doi.org/10.1128/AAC.02760-16
Huang W, Zhang Z, Ranade RM, Gillespie JR, Barros-Álvarez X, Creason SA, Shibata S, Verlinde CLMJ, Hol WGJ, Buckner FS, Fan E (2017) Optimization of a binding fragment targeting the “enlarged methionine pocket” leads to potent Trypanosoma brucei methionyl-tRNA synthetase inhibitors. Bioorg Med Chem Lett 27:2702–2707
Hussain T, Yogavel M, Sharma A (2015) Inhibition of protein synthesis and malaria parasite development by drug targeting of methionyl-tRNA synthetases. Antimicrob Agents Chemother 59:1856–1867
Ranade RM, Zhang Z, Gillespie JR, Shibata S, Verlinde CL, Hol WG, Fan E, Buckner FS (2015) Inhibitors of methionyl-tRNA synthetase have potent activity against Giardia intestinalis trophosoites. Antimicrob Agents Chemother 59:7128–7131
Jarvest RL, Berge JM, Berry V, Boyd HF, Brown MJ, Elder JS, Forrest AK, Fosberry AP, Gentry DR, Hibbs MJ, Jaworski DD, O'Hanlon PJ, Pope AJ, Rittenhouse S, Sheppard RJ, Slater-Radosti C, Worby A (2002) Nanomolar inhibitors of Staphylococcus aureus methionyl tRNA synthetase with potent antibacterial activity against gram-positive pathogens. J Med Chem 45:1959–1962
Eissa AG, Blaxland JA, Williams RO, Metwally KA, El-Adl SM, Lashine ESM, Baillie LW, Simons C (2016) Targeting methionyl-tRNA synthetase: design, synthesis and antibacterial activity against Clostridium difficile of novel 3-biaryl-N-benzylpropan-1-amine derivatives. J Enzyme Inhib Med Chem 31:1694–1697
Kumari M, Chandra S, Tiwari N, Subbarao N (2017) High throughput virtual screening to identify novel natural product inhibitors for methionyl-tRNA-synthetase of Brucella melitensis. Bioinformation 13:8–16
Robles S, Hu Y, Resto T, Dean F, Bullard JM (2017) Identification and characterization of a chemical compound that inhibits methionyl-tRNA synthetase from Pseudomonas aeruginosa. Curr Drug Discov Technol 14:156–168
Lee J, Kang SU, Kang MK, Chun MW, Jo YJ, Kwak JH, Kim S (1999) Methionyl adenylate analogues as inhibitors of methionyl-tRNA synthetase. Bioorg Med Chem Lett 9:1365–1370
Barros-Álvarez X, Turley S, Ranade RM, Gillespie JR, Duster NA, Verlinde CLMJ, Fan E, Buckner FS, Hol WGJ (2018) The crystal structure of the drug target Mycobacterium tuberculosis methionyl-tRNA synthetase in complex with a catalytic intermediate. Acta Crystallogr F 74:245–254
Wang W, Qin B, Wojdyla JA, Wang M, Gao X, Cui S (2018) Structural characterization of free-state and product-state Mycobacterium tuberculosis methionyl-tRNA synthetase reveals an induced-fit ligand-recognition mechanism. IUCrJ 5:478–490
Web-server PharmaGist. https://bioinfo3d.cs.tau.ac.il/PharmaGist/. Accessed 14 May 2019.
Dror O, Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ (2009) Novel approach for efficient pharmacophore-based virtual screening: method and applications. J Chem Inf Model 49:2333–2343
Discovery Studio Visualizer 4.0. https://www.3dsbiovia.com/products/collaborative-science/biovia-discovery-studio/visualization-download.php. Accessed 14 May 2019.
Starosyla SA, Volynets GP, Bdzhola VG, Yarmoluk SM (2016) Ukrainian certificate of registration of copyright for software “PharmDeveloper” № 70098.
Starosyla SA, Volynets GP, Protopopov MV, Bdzhola VG, Yarmoluk SM (2016) The development of algorithm for pharmacophore model optimization and rescoring of pharmacophore screening results. Ukr Bioorg Acta 1:24–34
National Center for Biotechnology Information. https://www.ncbi.nlm.nih.gov/. Accessed 14 May 2019.
Hess B, Kutzner C, van der Spoel D, Lindahl E (2008) GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4:435–447
Van der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC (2005) GROMACS: fast, flexible and free. J Comp Chem 26:1701–1719
Berendsen HJC, van der Spoel D, van Drunen R (1995) GROMACS: A message-passing parallel molecular dynamics implementation. Comp Phys Comm 91:43–56
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791
Pedretti A, Villa L, Vistoli G (2004) VEGA—an open platform to develop chemo-bio-informatics applications, using plug-in architecture and script programming. J Comput Aided Mol Des 18:167–173
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–252
Cogan EB, Birrell GB (1999) A robotics-based automated assay for inorganic and organic phosphates. Anal Biochem 271:29–35
Saenz-Méndez P, Eriksson LA (2018) Exploring polypharmacology in drug design. Methods Mol Biol 1824:229–243
Janardhan S, John L, Prasanthi M, Poroikov V, Narahari Sastry G (2017) A QSAR and molecular modelling study towards new lead finding: polypharmacological approach to Mycobacterium tuberculosis. SAR QSAR Environ Res 28:815–832
Fayaz SM, Rajanikant GK (2015) Ensembling and filtering: an effective and rapid in silico multitarget drug-design strategy to identify RIPK1 and RIPK3 inhibitors. J Mol Model 21:314
Domínguez JL, Fernández-Nieto F, Castro M, Catto M, Paleo MR, Porto S, Sardina FJ, Brea JM, Carotti A, Villaverde MC, Sussman F (2015) Computer-aided structure-based design of multitarget leads for Alzheimer’s disease. J Chem Inf Model 55:135–148
Arooj M, Sakkiah S, Gp G, Lee KW (2013) An innovative strategy for dual inhibitor design and its application in dual inhibition of human thymidylate synthase and dihydrofolate reductase enzymes. PLoS ONE 8:e60470
Chen Z, Han L, Xu M, Xu Y, Qian X (2013) Rationally designed multitarget anticancer agents. Curr Med Chem 20:1694–1714
Grisoni F, Merk D, Friedrich L, Schneider G (2019) Design of natural-product-inspired multitarget ligands by machine learning. ChemMedChem 14:1129–1134
Gudzera OI, Golub AG, Bdzhola VG, Volynets GP, Kovalenko OP, Boyarshin KS, Yaremchuk AD, Protopopov MV, Yarmoluk SM, Tukalo MA (2016) Identification of Mycobacterium tuberculosis leucyl-tRNA synthetase (LeuRS) inhibitors among the derivatives of 5-phenylamino-2H-[1,2,4]triazin-3-one. J Enzyme Inhib Med Chem 31:201–207
Acknowledgements
This work was supported by the Science and Technology Center in Ukraine (Contract No. 6258) and by the National Academy of Sciences of Ukraine (Contract No. 80–10/04–2019). Authors are grateful to Dr. Stephen Cusack and Dr. Andres Palencia (EMBL Grenoble Outstation, France) for the gift of plasmid encoding M. tuberculosis LeuRS. We also thank Prof. Vasyl Mel’nyk (National Institute of Phthisiology and Pulmonology named after F.G. Yanovsky of the NAMS of Ukraine, Kyiv, Ukraine) for providing the gene encoding M. tuberculosis MetRS.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Volynets, G.P., Starosyla, S.A., Rybak, M.Y. et al. Dual-targeted hit identification using pharmacophore screening. J Comput Aided Mol Des 33, 955–964 (2019). https://doi.org/10.1007/s10822-019-00245-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-019-00245-5