Abstract
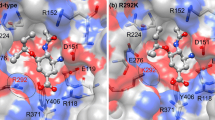
Influenza epidemics are responsible for an average of 3–5 millions of severe cases and up to 500,000 deaths around the world. One of flu pandemic types is influenza A(H1N1)pdm09 virus (pdm09H1N1). Oseltamivir is the antiviral drug used to treat influenza targeting at neuraminidase (NA) located on the viral surface. Influenza virus undergoes high mutation rates and leads to drug resistance, and thus the development of more efficient drugs is required. In the present study, all-atom molecular dynamics simulations were applied to understand the oseltamivir resistance caused by the single E119D and double E119D/H274Y mutations on NA. The obtained results in terms of binding free energy and intermolecular interactions in the ligand–protein interface showed that the oseltamivir could not be well accommodated in the binding pocket of both NA mutants and the 150-loop moves out from oseltamivir as an “open” state. A greater number of water molecules accessible to the binding pocket could disrupt the oseltamivir binding with NA target as seen be high mobility of oseltamivir at the active site. Additionally, our finding could guide to the design and development of novel NA inhibitor drugs.
Graphic abstract








Similar content being viewed by others
References
Arbeitskreis Blut UBBK (2009) Influenza virus. Transfus Med Hemother 36(1):32–39. https://doi.org/10.1159/000197314
Moorthy NSHN, Vasanthanathan P, Pratheepa V (2014) Viral M2 ion channel protein: a promising target for anti-influenza drug discovery. Mini Rev Med Chem 14(10):819–830. https://doi.org/10.2174/138955751410141020150822
Parra-Rojas C, Nguyen VK, Hernandez-Mejia G, Hernandez-Vargas EA (2018) Neuraminidase inhibitors in influenza treatment and prevention is it time to call it a day? Viruses 10(9):454. https://doi.org/10.3390/v10090454
Shen Z, Lou K, Wang W (2015) New small-molecule drug design strategies for fighting resistant influenza A. Acta Pharm Sin B 5(5):419–430. https://doi.org/10.1016/j.apsb.2015.07.006
Tan M, Wei C, Huang P, Fan Q, Quigley C, Xia M, Fang H, Zhang X, Zhong W, Klassen JS, Jiang X (2015) Tulane virus recognizes sialic acids as cellular receptors. Sci Rep 5:11784. https://doi.org/10.1038/srep11784
Dou D, Revol R, Östbye H, Wang H, Daniels R (2018) Influenza A virus cell entry, replication, virion assembly and movement. Front Immunol 9:1581–1581. https://doi.org/10.3389/fimmu.2018.01581
Hochgürtel M, Kroth H, Piecha D, Hofmann MW, Nicolau C, Krause S, Schaaf O, Sonnenmoser G, Eliseev AV (2002) Target-induced formation of neuraminidase inhibitors from in vitro virtual combinatorial libraries. Proc Natl Acad Sci USA 99(6):3382–3387. https://doi.org/10.1073/pnas.052703799
Kim CU, Chen X, Mendel DB (1999) Neuraminidase inhibitors as anti-influenza virus agents. Antivir Chem Chemother 10(4):141–154. https://doi.org/10.1177/095632029901000401
Samson M, Pizzorno A, Abed Y, Boivin G (2013) Influenza virus resistance to neuraminidase inhibitors. Antivir Res 98(2):174–185. https://doi.org/10.1016/j.antiviral.2013.03.014
Choi WS, Jeong JH, Kwon JJ, Ahn SJ, Lloren KKS, Kwon HI, Chae HB, Hwang J, Kim MH, Kim CJ, Webby RJ, Govorkova EA, Choi YK, Baek YH, Song MS (2018) Screening for neuraminidase inhibitor resistance markers among avian influenza viruses of the N4, N5, N6, and N8 neuraminidase subtypes. J Virol 92:1. https://doi.org/10.1128/jvi.01580-17
L'Huillier AG, Abed Y, Petty TJ, Cordey S, Thomas Y, Bouhy X, Schibler M, Simon A, Chalandon Y, van Delden C, Zdobnov E, Boquete-Suter P, Boivin G, Kaiser L (2015) E119D neuraminidase mutation conferring pan-resistance to neuraminidase inhibitors in an A(H1N1)pdm09 isolate from a stem-cell transplant recipient. J Infect Dis 212(11):1726–1734. https://doi.org/10.1093/infdis/jiv288
Baek YH, Song M-S, Lee E-Y, Kim Y-I, Kim E-H, Park S-J, Park KJ, Kwon H-I, Pascua PNQ, Lim G-J, Kim S, Yoon S-W, Kim MH, Webby RJ, Choi Y-K (2014) Profiling and characterization of influenza virus N1 strains potentially resistant to multiple neuraminidase inhibitors. J Virol 89(1):287–299. https://doi.org/10.1128/JVI.02485-14
Oh DY, Panozzo J, Vitesnik S, Farrukee R, Piedrafita D, Mosse J, Hurt AC (2018) Selection of multi-drug resistant influenza A and B viruses under zanamivir pressure and their replication fitness in ferrets. Antivir Ther 23(4):295–306. https://doi.org/10.3851/imp3135
Hamelin M-È, Baz M, Abed Y, Couture C, Joubert P, Beaulieu É, Bellerose N, Plante M, Mallett C, Schumer G, Kobinger GP, Boivin G (2010) Oseltamivir-resistant pandemic A/H1N1 virus is as virulent as its wild-type counterpart in mice and ferrets. PLOS Pathog 6(7):e1001015. https://doi.org/10.1371/journal.ppat.1001015
Vasudevan K, Veerappapillai S, Ramalingam R, Karuppasamy R (2012) Exploring the cause of oseltamivir resistance against mutant H274Y neuraminidase by molecular simulation approach. Appl Biochem Biotechnol 167:237–249. https://doi.org/10.1007/s12010-012-9687-7
Rungrotmongkol T, Malaisree M, Nunthaboot N, Sompornpisut P, Hannongbua S (2010) Molecular prediction of oseltamivir efficiency against probable influenza A (H1N1-2009) mutants: molecular modeling approach. Amino Acids 39(2):393–398. https://doi.org/10.1007/s00726-009-0452-3
Vavricka CJ, Li Q, Wu Y, Qi J, Wang M, Liu Y, Gao F, Liu J, Feng E, He J, Wang J, Liu H, Jiang H, Gao GF (2011) Structural and functional analysis of laninamivir and its octanoate prodrug reveals group specific mechanisms for influenza NA inhibition. PLOS Pathog 7(10):e1002249. https://doi.org/10.1371/journal.ppat.1002249
Case D, Betz RM, Cerutti DS, Cheatham T, Darden T, Duke R, Giese TJ, Gohlke H, Götz A, Homeyer N, Izadi S, Janowski P, Kaus J, Kovalenko A, Lee T-S, LeGrand S, Li P, Lin C, Luchko T, Kollman PA (2016) Amber, vol 16. University of California, San Francisco. https://doi.org/10.13140/RG.2.2.27958.70729
Udommaneethanakit T, Rungrotmongkol T, Bren U, Frecer V, Stanislav M (2009) Dynamic behavior of avian influenza A virus neuraminidase subtype H5N1 in complex with oseltamivir, zanamivir, peramivir, and their phosphonate analogues. J Chem Inf Mod 49(10):2323–2332. https://doi.org/10.1021/ci900277r
Perez A, MacCallum JL, Brini E, Simmerling C, Dill KA (2015) Grid-based backbone correction to the ff12SB protein force field for implicit-solvent simulations. J Chem Theory Comput 11(10):4770–4779. https://doi.org/10.1021/acs.jctc.5b00662
Olsson MHM, Søndergaard CR, Rostkowski M, Jensen JH (2011) PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions. J Chem Theory Comput 7(2):525–537. https://doi.org/10.1021/ct100578z
Mark P, Nilsson L (2001) Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J Phys Chem A 105(43):9954–9960. https://doi.org/10.1021/jp003020w
Sindhikara DJ, Yoshida N, Hirata F (2012) Placevent: an algorithm for prediction of explicit solvent atom distribution-application to HIV-1 protease and F-ATP synthase. J Comput Chem 33(18):1536–1543. https://doi.org/10.1002/jcc.22984
York DM, Darden TA, Pedersen LG (1993) The effect of long-range electrostatic interactions in simulations of macromolecular crystals: a comparison of the Ewald and truncated list methods. J Chem Phys 99:8345–8348. https://doi.org/10.1063/1.465608
Phanich J, Rungrotmongkol T, Kungwan N, Hannongbua S (2016) Role of R292K mutation in influenza H7N9 neuraminidase toward oseltamivir susceptibility: MD and MM/PB(GB)SA study. J Comput Aided Mol Des 30:917–926. https://doi.org/10.1007/s10822-016-9981-5
Hou T, Wang J, Li Y, Wang W (2011) Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J Chem Inf Model 51(1):69–82. https://doi.org/10.1021/ci100275a
Best RB, Hummer G, Eaton WA (2013) Native contacts determine protein folding mechanisms in atomistic simulations. Proc Natl Acad Sci 110(44):17874. https://doi.org/10.1073/pnas.1311599110
Ali SA, Hassan MI, Islam A, Ahmad F (2014) A review of methods available to estimate solvent-accessible surface areas of soluble proteins in the folded and unfolded states. Curr Protein Peptide Sci 15(5):456–476
Roe DR, Cheatham TE 3rd (2013) PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J Chem Theory Comput 9(7):3084–3095. https://doi.org/10.1021/ct400341p
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J Comput Chem 25(13):1605–1612. https://doi.org/10.1002/jcc.20084
Sirikantaramas S, Meeprasert A, Rungrotmongkol T, Fuji H, Hoshino T, Sudo H, Yamazaki M, Saito K (2015) Structural insight of DNA topoisomerases I from camptothecin-producing plants revealed by molecular dynamics simulations. Phytochemistry 113:50–56. https://doi.org/10.1016/j.phytochem.2015.02.012
Panman W, Nutho B, Chamni S, Dokmaisrijan S, Kungwan N, Rungrotmongkol T (2018) Computational screening of fatty acid synthase inhibitors against thioesterase domain. J Biomol Struct Dyn 36(15):4114–4125. https://doi.org/10.1080/07391102.2017.1408496
Genheden S, Ryde U (2015) The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin Drug Discov 10(5):449–461. https://doi.org/10.1517/17460441.2015.1032936
Peddi SR, Sivan SK, Manga V (2018) Molecular dynamics and MM/GBSA-integrated protocol probing the correlation between biological activities and binding free energies of HIV-1 TAR RNA inhibitors. J Biomol Struct Dyn 36(2):486–503. https://doi.org/10.1080/07391102.2017.1281762
Rizzo RC, Udier-Blagovic M, Wang DP, Watkins EK, Kroeger Smith MB, Smith RH Jr, Tirado-Rives J, Jorgensen WL (2002) Prediction of activity for nonnucleoside inhibitors with HIV-1 reverse transcriptase based on Monte Carlo simulations. J Med Chem 45(14):2970–2987. https://doi.org/10.1021/jm010580q
Wang J, Morin P, Wang W, Kollman PA (2001) Use of MM-PBSA in reproducing the binding free energies to HIV-1 RT of TIBO derivatives and predicting the binding mode to HIV-1 RT of efavirenz by docking and MM-PBSA. J Am Chem Soc 123(22):5221–5230. https://doi.org/10.1021/ja003834q
Rungrotmongkol T, Udommaneethanakit T, Malaisree M, Nunthaboot N, Intharathep P, Sompornpisut P, Hannongbua S (2009) How does each substituent functional group of oseltamivir lose its activity against virulent H5N1 influenza mutants? Biophys chem 145(1):29–36. https://doi.org/10.1016/j.bpc.2009.08.006
Phanich J, Rungrotmongkol T, Sindhikara D, Phongphanphanee S, Yoshida N, Hirata F, Kungwan N, Hannongbua S (2016) A 3D-RISM/RISM study of the oseltamivir binding efficiency with the wild-type and resistance-associated mutant forms of the viral influenza B neuraminidase. Protein Sci 25(1):147–158. https://doi.org/10.1002/pro.2718
Wu Y, Gao F, Qi J, Bi Y, Fu L, Mohan S, Chen Y, Li X, Pinto BM, Vavricka CJ, Tien P, Gao GF (2016) Resistance to mutant group 2 influenza virus neuraminidases of an oseltamivir-zanamivir hybrid inhibitor. J Virol 90(23):10693. https://doi.org/10.1128/JVI.01703-16
Van der Vries E, Collins PJ, Vachieri S, Xiong X, Liu J, Walker PA, Haire LF, Hay AJ, Schutten M, Osterhaus ADME, Martin SR, Boucher C, Skehel JJ, Gamblin SJ (2012) H1N1 2009 pandemic influenza virus: resistance of the I223R Neuraminidase mutant explained by kinetic and structural analysis. PLoS Pathog 8:e1002914. https://doi.org/10.1371/journal.ppat.1002914
Wu Y, Bi Y, Vavricka C, Sun X, Zhang Y, Gao F, Zhao M, Xiao H, Qin C-F, He J, Wenjun L, Yan J, Qi J, Gao GF (2013) Characterization of two distinct neuraminidases from avian-origin human-infecting H7N9 influenza viruses. Cell Res 23:1347. https://doi.org/10.1038/cr.2013.144
Palmer J, Dobrovolny HM, Beauchemin CAA (2017) The in vivo efficacy of neuraminidase inhibitors cannot be determined from the decay rates of influenza viral titers observed in treated patients. Sci Rep 7:40210–40210. https://doi.org/10.1038/srep40210
Yen HL, Hoffmann E, Taylor G, Scholtissek C, Monto AS, Webster RG, Govorkova EA (2006) Importance of neuraminidase active-site residues to the neuraminidase inhibitor resistance of influenza viruses. J Virol 80(17):8787–8795. https://doi.org/10.1128/jvi.00477-06
Mandal RS, Panda S, Das S (2018) In silico prediction of drug resistance due to S247R mutation of Influenza H1N1 neuraminidase protein. J Biomol Struct Dyn 36(4):966–980. https://doi.org/10.1080/07391102.2017.1305295
Wu Y, Qin G, Gao F, Liu Y, Vavricka CJ, Qi J, Jiang H, Yu K, Gao GF (2013) Induced opening of influenza virus neuraminidase N2 150-loop suggests an important role in inhibitor binding. Sci Rep 3:1551. https://doi.org/10.1038/srep01551
Du Q-S, Wang S-Q, Chou K-C (2007) Analogue inhibitors by modifying oseltamivir based on the crystal neuraminidase structure for treating drug-resistant H5N1 virus. Biochem Biophys Res Commun 362(2):525–531. https://doi.org/10.1016/j.bbrc.2007.08.025
D'Souza C, Kanyalkar M, Joshi M, Coutinho E, Srivastava S (2009) Search for novel neuraminidase inhibitors: design, synthesis and interaction of oseltamivir derivatives with model membrane using docking, NMR and DSC methods. Biochim Biophys Acta 1788(9):1740–1751. https://doi.org/10.1016/j.bbamem.2009.04.014
Acknowledgements
This research was supported by the Research Chair Grant, the National Science and Technology Development Agency (NSTDA), Thailand. C.H. thanks Science Achievement Scholarship of Thailand and Dr.Bodee Nutho for resolving some technical problems. K.T. thanks Chulalongkorn University for supporting a research visit. The Structural and Computational Biology Research Unit is acknowledged for facility and computing resources.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hanpaibool, C., Leelawiwat, M., Takahashi, K. et al. Source of oseltamivir resistance due to single E119D and double E119D/H274Y mutations in pdm09H1N1 influenza neuraminidase. J Comput Aided Mol Des 34, 27–37 (2020). https://doi.org/10.1007/s10822-019-00251-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-019-00251-7