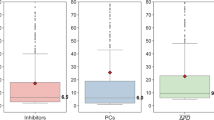
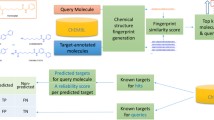
Abstract
Small molecules with multi-target activity, also termed promiscuous compounds, are increasingly considered for pharmaceutical applications. The use of promiscuous chemical entities represents a departure from the compound specificity paradigm, one of the pillars of modern drug discovery. The popularity of promiscuous compounds is due to the concept of polypharmacology; another more recent drug discovery paradigm. It refers to insights that the efficacy of drugs often depends on interactions with multiple targets. Views concerning the extent to which small molecules might form well-defined interactions with multiple targets often differ, but comprehensive experimental investigations of promiscuity are currently rare. On the other hand, large volumes of active compounds and experimental measurements are becoming available and enable data-driven analyses of compound selectivity versus promiscuity. In this perspective, we discuss computational methods and data structures designed for promiscuity analysis. In addition, findings from large-scale exploration of activity profiles of inhibitors covering the human kinome are summarized. Although many kinase inhibitors are expected to be promiscuous, they are frequently found to be selective, which provides opportunities for target-directed drug discovery (rather than polypharmacology). We also discuss that machine learning yields evidence for the existence of structure–promiscuity relationships.




Similar content being viewed by others
References
Hu Y, Bajorath J (2013) Compound promiscuity: what can we learn from current data? Drug Discov Today 18:644–650
Hu Y, Bajorath J (2017) Entering the ‘big data’ era in medicinal chemistry: molecular promiscuity analysis revisited. Future Sci OA 3:FSO179
McGovern SL, Caselli E, Grigorieff N, Shoichet BK (2002) A common mechanism underlying promiscuous inhibitors from virtual and high-throughput screening. J Med Chem 45:1712–1722
Feng BY, Shelat A, Doman TN, Guy RK, Shoichet BK (2005) High-throughput assays for promiscuous inhibitors. Nat Chem Biol 1:146–148
Shoichet BK (2006) Screening in a spirit haunted world. Drug Discov Today 11:607–615
Baell JB, Holloway GA (2010) New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J Med Chem 53:2719–2740
Baell J, Walters MA (2014) Chemistry: chemical con artists foil drug discovery. Nature 513:481–483
Anighoro A, Bajorath J, Rastelli G (2014) Polypharmacology: challenges and opportunities in drug discovery. J Med Chem 57:7874–7887
Proschak E, Stark H, Merk D (2019) Polypharmacology by design: a medicinal chemist’s perspective on multitargeting compounds. J Med Chem 62:420–444
Bolognesi ML (2019) Harnessing polypharmacology with medicinal chemistry. ACS Med Chem Lett 10:273–275
Mei Y, Yang B (2018) Rational application of drug promiscuity in medicinal chemistry. Future Med Chem 10:1835–1851
Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, Chan KW, Ciceri P, Davis MI, Edeen PT, Faraoni R, Floyd M, Hunt JP, Lockhart DJ, Milanov ZV, Morrison MJ, Pallares G, Patel HK, Pritchard S, Wodicka LM, Zarrinkar PP (2008) A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol 26:127–132
Anastassiadis T, Deacon SW, Devarajan K, Ma H, Peterson JR (2011) Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nat Biotechnol 29:1039–1045
Elkins JM, Fedele V, Szklarz M, Abdul Azeez KR, Salah E, Mikolajczyk J, Romanov S, Sepetov N, Huang XP, Roth BL, Al Haj Zen A, Fourches D, Muratov E, Tropsha A, Morris J, Teicher BA, Kunkel M, Polley E, Lackey KE, Atkinson FL, Overington JP, Bamborough P, Müller S, Price DJ, Willson TM, Drewry DH, Knapp S, Zuercher WJ (2016) Comprehensive characterization of the published kinase inhibitor set. Nat Biotechnol 34:95–103
Klaeger S, Heinzlmeir S, Wilhelm M, Polzer H, Vick B, Koenig PA, Reinecke M, Ruprecht B, Petzoldt S, Meng C, Zecha J, Reiter K, Qiao H, Helm D, Koch H, Schoof M, Canevari G, Casale E, Depaolini SR, Feuchtinger A, Wu Z, Schmidt T, Rueckert L, Becker W, Huenges J, Garz AK, Gohlke BO, Zolg DP, Kayser G, Vooder T, Preissner R, Hahne H, Tõnisson N, Kramer K, Götze K, Bassermann F, Schlegl J, Ehrlich HC, Aiche S, Walch A, Greif PA, Schneider S, Felder ER, Ruland J, Médard G, Jeremias I, Spiekermann K, Kuster B (2017) The target landscape of clinical kinase drugs. Science 358:eaan4368
Hu Y, Bajorath J (2013) High-resolution view of compound promiscuity. F1000Research 2:e144
Jasial S, Hu Y, Bajorath J (2016) Determining the degree of promiscuity of extensively assayed compounds. PLoS ONE 11:e0153873
Stumpfe D, Tinivella A, Rastelli G, Bajorath J (2017) Promiscuity of inhibitors of human protein kinases at varying data confidence levels and test frequencies. RSC Adv 7:41265–41271
Keiser MJ, Setola V, Irwin JJ, Laggner C, Abbas AI, Hufeisen SJ, Jensen NH, Kuijer MB, Matos RC, Tran TB, Whaley R, Glennon RA, Hert J, Thomas KLH, Edwards DD, Shoichet BK, Roth BL (2009) Predicting new molecular targets for known drugs. Nature 462:175–181
Keiser MJ, Roth BL, Armbruster BN, Ernsberger P, Irwin JJ, Shoichet BK (2007) Relating protein pharmacology by ligand chemistry. Nat Biotechnol 25:197–206
Wang L, Ma C, Wipf P, Liu H, Su W, Xie X-Q (2013) TargetHunter: an in silico target identification tool for predicting therapeutic potential of small organic molecules based on chemogenomic database. AAPS J 15:395–406
Awale M, Reymond J-L (2017) The polypharmacology browser: a web-based multi-fingerprint target prediction tool using ChEMBL bioactivity data. J Cheminform 9:e11
Jasial S, Gilberg E, Blaschke T, Bajorath J (2018) Machine learning distinguishes with high accuracy between pan-assay interference compounds that are promiscuous or represent dark chemical matter. J Med Chem 61:10255–10264
Blaschke T, Miljković F, Bajorath J (2019) Prediction of different classes of promiscuous and nonpromiscuous compounds using machine learning and nearest neighbor analysis. ACS Omega 4:6883–6890
Miljković F, Kunimoto R, Bajorath J (2017) Identifying relationships between unrelated pharmaceutical target proteins on the basis of shared active compounds. Future Sci OA 3:FSO212
Reker D, Bernardes GJL, Rodrigues T (2019) Computational advances in combating colloidal aggregation in drug discovery. Nat Chem 11:402–418
Dantas RF, Evangelista TCS, Neves BJ, Senger MR, Andrade CH, Ferreira SB, Silva-Junior FP (2019) Dealing with frequent hitters in drug discovery: a multidisciplinary view on the issue of filtering compounds on biological screenings. Expert Opin Drug Discov. https://doi.org/10.1080/17460441.2019.1654453
Haupt JV, Daminelli S, Schroeder M (2013) Drug promiscuity in PDB: protein binding site similarity is key. PLoS ONE 8:e65894
Pinzi L, Caporuscio F, Rastelli G (2018) Selection of protein conformations for structure-based polypharmacology studies. Drug Discov Today 23:1889–1896
Gilberg E, Stumpfe D, Bajorath J (2018) X-ray structure-based identification of compounds with activity against targets from different families and generation of templates for multitarget ligand design. ACS Omega 3:106–111
Gilberg E, Gütschow M, Bajorath J (2019) Promiscuous ligands from experimentally determined structures, binding conformations, and protein family-dependent interaction hotspots. ACS Omega 4:1729–1737
Gilberg E, Bajorath J (2019) Recent progress in structure-based evaluation of compound promiscuity. ACS Omega 4:2758–2765
Mestres J, Gregori-Puigjané E, Valverde S, Solé RV (2008) Data completeness—the Achilles heel of drug-target networks. Nat Biotechnol 26:983–984
Mestres J, Gregori-Puigjané E, Valverde S, Solé RV (2009) The topology of drug-target interaction networks: implicit dependence on drug properties and target families. Mol BioSyst 5:1051–1057
Maggiora GM (2006) On outliers and activity cliffs—why QSAR often disappoints. J Chem Inf Model 46:1535
Stumpfe D, Bajorath J (2012) Exploring activity cliffs in medicinal chemistry. J Med Chem 55:2932–2942
Dimova D, Bajorath J (2018) Rationalizing promiscuity cliffs. ChemMedChem 13:490–494
Kenny PW, Sadowski J (2005) Structure modification in chemical databases. In: Oprea TI (ed) Chemoinformatics in drug discovery. Wiley-VCH, Weinheim, pp 271–285
Hu Y, Jasial S, Gilberg E, Bajorath J (2017) Structure-promiscuity relationship puzzles—extensively assayed analogs with large differences in target annotations. AAPS J 19:856–864
Dimova D, Gilberg E, Bajorath J (2017) Identification and analysis of promiscuity cliffs formed by bioactive compounds and experimental implications. RSC Adv 7:58–66
Miljković F, Bajorath J (2018) Computational analysis of kinase inhibitors identifies promiscuity cliffs across the human kinome. ACS Omega 3:17295–17308
Miljković F, Vogt M, Bajorath J (2019) Systematic computational identification of promiscuity cliff pathways formed by inhibitors of the human kinome. J Comput Aided Mol Des 33:559–572
Dijkstra EW (1959) A note on two problems in connexion with graphs. Numer Math 1:269–271
Knight ZA, Lin H, Shokat KM (2010) Targeting the cancer kinome through polypharmacology. Nat Rev Cancer 10:130–137
Miljković F, Bajorath J (2019) Data structures for compound promiscuity analysis: promiscuity cliffs, pathways and promiscuity hubs formed by inhibitors of the human kinome. Fut Sci OA 5:FSO404
Cohen P (2002) Protein kinases—the major drug targets of the twenty-first century? Nat Rev Drug Discovery 1:309–315
Simmons DL (2013) Targeting kinases: a new approach to treating inflammatory rheumatic diseases. Curr Opin Pharmacol 13:426–434
Laufer S, Bajorath J (2014) New frontiers in kinases: second generation inhibitors. J Med Chem 57:2167–2168
Gavrin LK, Saiah E (2013) Approaches to discover non-ATP site kinase inhibitors. Med Chem Commun 4:41–51
Zhao Z, Wu H, Wang L, Liu Y, Knapp S, Liu Q, Gray NS (2014) Exploration of type II binding mode: a privileged approach for kinase inhibitor focused drug discovery? ACS Chem Biol 9:1230–1241
Hu Y, Furtmann N, Bajorath J (2015) Current compound coverage of the kinome. J Med Chem 58:30–40
Miljković F, Rodríguez-Pérez R, Bajorath J (2019) Machine learning models for accurate prediction of kinase inhibitors with different binding modes. J Med Chem. https://doi.org/10.1021/acs.jmedchem.9b00867
Miljković F, Bajorath J (2018) Exploring selectivity of multikinase inhibitors across the human kinome. ACS Omega 3:1147–1153
Miljković F, Bajorath J (2018) Reconciling selectivity trends from a comprehensive kinase inhibitor profiling campaign with known activity data. ACS Omega 3:3113–3119
Levitzki A (2013) Tyrosine kinase inhibitors: views of selectivity, sensitivity, and clinical performance. Annu Rev Pharmacol Toxicol 53:161–185
Wu P, Dinér P, Bunch L (2018) The screening and design of allosteric kinase inhibitors. In: Ward RA, Goldberg FW (eds) Kinase drug discovery: modern approaches. RSC, Cambridge, pp 34–60
Miljković F, Bajorath J (2018) Data-driven exploration of selectivity and off-target activities of designated chemical probes. Molecules 23:e2434
Rodríguez-Pérez R, Bajorath J (2019) Multitask machine learning for classifying highly and weakly potent kinase inhibitors. ACS Omega 4:4367–4375
Li X, Li Z, Wu X, Xiong Z, Yang T, Fu Z, Liu X, Tan X, Zhong F, Wan X, Wang D, Ding X, Yang R, Hou H, Li C, Liu H, Chen K, Jiang H, Zheng M (2019) Deep learning enhancing kinome-wide polypharmacology profiling: model construction and experiment validation. J Med Chem. https://doi.org/10.1021/acs.jmedchem.9b00855
Li Z, Li X, Liu X, Fu Z, Xiong Z, Wu X, Tan X, Zhao J, Zhong F, Wan X, Luo X, Chen K, Jiang H, Zheng M (2019) KinomeX: a web application for predicting kinome-wide polypharmacology effect on small molecules. Bioinformatics. https://doi.org/10.1093/bioinformatics/btz519
Breiman L (2001) Random forests. Mach Learn 45:5–32
Joachims T (1999) Making large-scale SVM learning practical. In: Schölkopf B, Burges CJC, Smola AJ (eds) Advances in kernel methods: support vector learning. MIT Press, Cambridge, pp 169–184
Goodfellow I, Bengio Y, Courville A (2016) Deep learning. MIT Press, Cambridge
Duvenaud D, Maclaurin D, Aguilera-Iparraguirre J, Gomez-Bombarelli R, Hirzel T, Aspuru-Guzik A, Adams RP (2015) Convolutional networks on graph for learning molecular fingerprints. Neural Inf Proc Sys 28:2224–2232
Balfer J, Bajorath J (2015) Visualization and interpretation of support vector machine activity predictions. J Chem Inf Model 55:1136–1147
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Miljković, F., Bajorath, J. Data structures for computational compound promiscuity analysis and exemplary applications to inhibitors of the human kinome. J Comput Aided Mol Des 34, 1–10 (2020). https://doi.org/10.1007/s10822-019-00266-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-019-00266-0