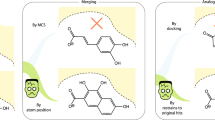
Abstract
Optimization in medicinal chemistry often involves designing replacements for a section of a molecule which aim to retain potency while improving other properties of the compound. In this study, we perform a retrospective analysis using a number of computational methods to identify active side chains amongst a pool of random decoy side chains, mimicking a similar procedure that might be undertaken in a real medicinal chemistry project. We constructed a dataset derived from public ChEMBL and PDB data by identifying all ChEMBL assays where at least one of the compounds tested has also been co-crystallized in the PDB. Additionally, we required that there be at least ten active compounds tested in the same ChEMBL assay that are matched molecular pairs to the crystallized ligand. Using the compiled dataset consisting of sets of compounds from 402 assays, we have tested a number of methods for scoring side chains including Spark, a bioisostere replacement tool from Cresset, molecular docking using Glide from Schrodinger, docking with Smina, as well as other methods. In this work, we present a comparison of the performance of these methods in discriminating active side chains from decoys as well as recommendations for circumstances when different methods should be used.










Similar content being viewed by others
References
Ripphausen P, Nisius B, Peltason L, Bajorath J (2010) Quo vadis, virtual screening? A comprehensive survey of prospective applications. J Med Chem 53(24):8461–8467. doi:https://doi.org/10.1021/jm101020z
Truchon JF, Bayly CI (2007) Evaluating virtual screening methods: good and bad metrics for the “early recognition” problem. J Chem Inf Model 47(2):488–508. doi:https://doi.org/10.1021/ci600426e
Huang N, Shoichet BK, Irwin JJ (2006) Benchmarking sets for molecular docking. J Med Chem 49(23):6789–6801. doi:https://doi.org/10.1021/jm0608356
Venkatraman V, Perez-Nueno VI, Mavridis L, Ritchie DW (2010) Comprehensive comparison of ligand-based virtual screening tools against the DUD data set reveals limitations of current 3D methods. J Chem Inf Model 50(12):2079–2093. doi:https://doi.org/10.1021/ci100263p
Riniker S, Landrum GA (2013) Open-source platform to benchmark fingerprints for ligand-based virtual screening. J Cheminform 5(1):26. doi:https://doi.org/10.1186/1758-2946-5-26
Leach AG, Jones HD, Cosgrove DA, Kenny PW, Ruston L, MacFaul P, Wood JM, Colclough N, Law B (2006) Matched molecular pairs as a guide in the optimization of pharmaceutical properties; a study of aqueous solubility, plasma protein binding and oral exposure. J Med Chem 49(23):6672–6682. doi:https://doi.org/10.1021/jm0605233
Griffen E, Leach AG, Robb GR, Warner DJ (2011) Matched molecular pairs as a medicinal chemistry tool. J Med Chem 54(22):7739–7750. doi:https://doi.org/10.1021/jm200452d
Bento AP, Gaulton A, Hersey A, Bellis LJ, Chambers J, Davies M, Kruger FA, Light Y, Mak L, McGlinchey S, Nowotka M, Papadatos G, Santos R, Overington JP (2014) The ChEMBL bioactivity database: an update. Nucleic Acids Res 42(Database issue):D1083–D1090. doi:https://doi.org/10.1093/nar/gkt1031
Gaulton A, Bellis LJ, Bento AP, Chambers J, Davies M, Hersey A, Light Y, McGlinchey S, Michalovich D, Al-Lazikani B, Overington JP (2012) ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res 40(Database issue):D1100–D1107. doi:https://doi.org/10.1093/nar/gkr777
Gaulton A, Hersey A, Nowotka M, Bento AP, Chambers J, Mendez D, Mutowo P, Atkinson F, Bellis LJ, Cibrian-Uhalte E, Davies M, Dedman N, Karlsson A, Magarinos MP, Overington JP, Papadatos G, Smit I, Leach AR (2017) The ChEMBL database in 2017. Nucleic Acids Res 45(D1):D945–D954. doi:https://doi.org/10.1093/nar/gkw1074
Erl Wood Cheminformatics nodes for KNIME (2017)
Wagener M, Lommerse JP (2006) The quest for bioisosteric replacements. J Chem Inf Model 46(2):677–685. doi:https://doi.org/10.1021/ci0503964
Hussain J, Rea C (2010) Computationally efficient algorithm to identify matched molecular pairs (MMPs) in large data sets. J Chem Inf Model 50(3):339–348. doi:https://doi.org/10.1021/ci900450m
Papadatos G, Alkarouri M, Gillet VJ, Willett P, Kadirkamanathan V, Luscombe CN, Bravi G, Richmond NJ, Pickett SD, Hussain J, Pritchard JM, Cooper AW, Macdonald SJ (2010) Lead optimization using matched molecular pairs: inclusion of contextual information for enhanced prediction of HERG inhibition, solubility, and lipophilicity. J Chem Inf Model 50(10):1872–1886. doi:https://doi.org/10.1021/ci100258p
Mysinger MM, Carchia M, Irwin JJ, Shoichet BK (2012) Directory of useful decoys, enhanced (DUD-E): better ligands and decoys for better benchmarking. J Med Chem 55(14):6582–6594. doi:https://doi.org/10.1021/jm300687e
Landrum G RDKit: Open Source cheminformatics
Koes DR, Baumgartner MP, Camacho CJ (2013) Lessons learned in empirical scoring with smina from the CSAR 2011 benchmarking exercise. J Chem Inf Model 53(8):1893–1904. doi:https://doi.org/10.1021/ci300604z
Smina Apr 2 2016 build. https://sourceforge.net/projects/smina/
Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS (2004) Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem 47(7):1739–1749. doi:https://doi.org/10.1021/jm0306430
Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, Banks JL (2004) Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J Med Chem 47(7):1750–1759. doi:https://doi.org/10.1021/jm030644s
Schrödinger Release 2017-2. Protein Preparation Wizard; Epik, Schrödinger LLC, New York NY, 2016; Impact, Schrödinger, LLC, New York, NY, 2016; Prime, Schrödinger, LLC, New York, NY, 2016.
Sastry GM, Adzhigirey M, Day T, Annabhimoju R, Sherman W (2013) Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des 27(3):221–234. https://doi.org/10.1007/s10822-013-9644-8
Spark, 10.4.0, Cresset®, Litlington, Cambridgeshire, UK; http://www.cresset-group.com/spark/;
Cheeseright T, Mackey M, Rose S, Vinter A (2006) Molecular field extrema as descriptors of biological activity: definition and validation. J Chem Inf Model 46(2):665–676. https://doi.org/10.1021/ci050357s
Watts KS, Dalal P, Murphy RB, Sherman W, Friesner RA, Shelley JC (2010) ConfGen: a conformational search method for efficient generation of bioactive conformers. J Chem Inf Model 50(4):534–546. https://doi.org/10.1021/ci100015j
Daylight Theory Manual http://www.daylight.com/dayhtml/doc/theory/index.pdf
O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR (2011) Open babel: an open chemical toolbox. J Cheminform 3:33. https://doi.org/10.1186/1758-2946-3-33
Koes DR. https://github.com/dkoes/asacalc/blob/master/asacalc.cpp. Accessed 2016
Bemis GW, Murcko MA (1996) The properties of known drugs. 1. Molecular frameworks. J Med Chem 39(15):2887–2893. doi:https://doi.org/10.1021/jm9602928
Erickson JA, Jalaie M, Robertson DH, Lewis RA, Vieth M (2004) Lessons in molecular recognition: the effects of ligand and protein flexibility on molecular docking accuracy. J Med Chem 47(1):45–55. doi:https://doi.org/10.1021/jm030209y
Abel R, Wang L, Harder ED, Berne BJ, Friesner RA (2017) Advancing drug discovery through enhanced free energy calculations. Acc Chem Res 50(7):1625–1632. https://doi.org/10.1021/acs.accounts.7b00083
Acknowledgements
The research leading to these results has received funding from the European Union’s Seventh Framework Programme (FP7/2007–2013) under grant agreement n°612347. The authors wish to acknowledge Lewis Vidler for constructive discussion and feedback and Jeremy Desaphy for the prepared PDB structures.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Baumgartner, M.P., Evans, D.A. Side chain virtual screening of matched molecular pairs: a PDB-wide and ChEMBL-wide analysis. J Comput Aided Mol Des 34, 953–963 (2020). https://doi.org/10.1007/s10822-020-00313-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-020-00313-1