Abstract
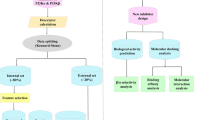
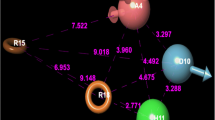
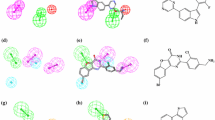
Herein, the LASSBio Chemical Library is presented as a valuable source of compounds for screening to identify hits suitable for subsequent hit-to-lead optimization stages. A feature of the LASSBio Chemical Library worth highlighting is the fact that it is a smart library designed by medicinal chemists with pharmacological activity as the main priority. The great majority of the compounds part of this library have shown in vivo activity in animal models, which is an indication that they possess overall favorable bioavailability properties and, hence, adequate pharmacokinetic profiles. This, in turn, is supported by the fact that approximately 85% of the compounds are compliant with Lipinski’s rule of five and ca. 95% are compliant with Veber’s rules, two important guidelines for oral bioavailability. In this work it is presented a virtual screening methodology combining a pharmacophore-based model and an empirical Gibbs free energy-based model for the ligand–protein interaction to explore the LASSBio Chemical Library as a source of new hits for the inhibition of the phosphatidylinositol 4-kinase IIIβ (PI4KIIIβ) enzyme, which is related to the development of viral infections (including enteroviruses, SARS coronavirus, and hepatitis C virus), cancers and neurological diseases. The approach resulted in the identification of two hits, LASSBio-1799 (7) and LASSBio-1814 (10), which inhibited the target enzyme with IC50 values of 3.66 μM and IC50 and 6.09 μM, respectively. This study also enabled the determination of the structural requirements for interactions with the active site of PI4KIIIβ, demonstrating the importance of both acceptor and donor hydrogen bonding groups for forming interactions with binding site residues Val598 and Lys549.




Similar content being viewed by others
References
Holenz J, Stoy P (2019) Advances in lead generation. Bioorg Med Chem Lett 29:517–524. https://doi.org/10.1016/j.bmcl.2018.12.001
Wermuth CG, Aladous D, Raboisson P, Rognan D (2015) The Practice of Medicinal Chemistry, 4th edn. London, Elsevier
Hersey A, Chambers J, Bellis L et al (2015) Chemical databases: curation or integration by user-defined equivalence? Drug Discov Today Technol 14:17–24. https://doi.org/10.1016/j.ddtec.2015.01.005
Walters WP (2019) Virtual chemical libraries. J Med Chem 62:1116–1124. https://doi.org/10.1021/acs.jmedchem.8b01048
Follmann M, Briem H, Steinmeyer A et al (2019) An approach towards enhancement of a screening library: the Next Generation Library Initiative (NGLI) at Bayer: against all odds? Drug Discov Today 24:668–672. https://doi.org/10.1016/j.drudis.2018.12.003
Vieth M, Siegel MG, Higgs RE et al (2004) Characteristic physical properties and structural fragments of marketed oral drugs. J Med Chem 47:224–232. https://doi.org/10.1021/jm030267j
Ohno K, Nagahara Y, Tsunoyama K, Orita M (2010) Are there differences between launched drugs, clinical candidates, and commercially available compounds? J Chem Inf Model 50:815–821. https://doi.org/10.1021/ci100023s
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (1997) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings 1PII of original article: S0169–409X(96), 00423–1. The article was originally published in Advanced Drug Delivery Reviews 23 (1997). Adv Drug Deliv Rev 23:3–26. https://doi.org/10.1016/S0169-409X(00)00129-0
Veber DF, Johnson SR, Cheng H-Y et al (2002) Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem 45:2615–2623. https://doi.org/10.1021/jm020017n
Wang Y, Xiao J, Suzek TO et al (2009) PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res 37:W623–W633. https://doi.org/10.1093/nar/gkp456
Irwin JJ, Shoichet BK (2005) ZINC: a free database of commercially available compounds for virtual screening. J Chem Inf Model 45:177–182. https://doi.org/10.1021/ci049714+
Prestwick Chemical. https://www.prestwickchemical.com/libraries-screening-lib-pcl.html. Accessed 2 Jul 2019
Wood A, Armour D (2005) The discovery of the CCR5 receptor antagonist, UK-427,857, a new agent for the treatment of HIV infection and AIDS. In: King FD (ed) Progress in medicinal chemistry. Elsevier, London, pp 239–271
LASSBio Chemical Library. https://www.lassbiochemicallibrary.com. Accessed 2 Jul 2019
Richard AW, Frederick G (2011) Kinase drug discovery, 1st edn. Royal Society of Chemistry, Cambridge
Borawski J, Troke P, Puyang X et al (2009) Class III phosphatidylinositol 4-kinase alpha and beta are novel host factor regulators of hepatitis C virus replication. J Virol 83:10058–10074. https://doi.org/10.1128/JVI.02418-08
Li J, Lu Y, Zhang J et al (2010) PI4KIIα is a novel regulator of tumor growth by its action on angiogenesis and HIF-1α regulation. Oncogene 29:2550–2559. https://doi.org/10.1038/onc.2010.14
Moorhead AM, Jung JY, Smirnov A et al (2010) Multiple host proteins that function in phosphatidylinositol-4-phosphate metabolism are recruited to the chlamydial inclusion. Infect Immun 78:1990–2007. https://doi.org/10.1128/IAI.01340-09
Delang L, Paeshuyse J, Neyts J (2012) The role of phosphatidylinositol 4-kinases and phosphatidylinositol 4-phosphate during viral replication. Biochem Pharmacol 84:1400–1408. https://doi.org/10.1016/j.bcp.2012.07.034
Clayton EL, Minogue S, Waugh MG (2013) Phosphatidylinositol 4-kinases and PI4P metabolism in the nervous system: roles in psychiatric and neurological diseases. Mol Neurobiol 47:361–372. https://doi.org/10.1007/s12035-012-8358-6
Waugh MG (2012) Phosphatidylinositol 4-kinases, phosphatidylinositol 4-phosphate and cancer. Cancer Lett 325:125–131. https://doi.org/10.1016/j.canlet.2012.06.009
Altan-Bonnet N, Balla T (2012) Phosphatidylinositol 4-kinases: hostages harnessed to build panviral replication platforms. Trends Biochem Sci 37:293–302. https://doi.org/10.1016/j.tibs.2012.03.004
Yang N, Ma P, Lang J et al (2012) Phosphatidylinositol 4-kinase IIIβ is required for severe acute respiratory syndrome coronavirus spike-mediated cell entry. J Biol Chem 287:8457–8467. https://doi.org/10.1074/jbc.M111.312561
Boura E, Nencka R (2015) Phosphatidylinositol 4-kinases: function, structure, and inhibition. Exp Cell Res 337:136–145. https://doi.org/10.1016/j.yexcr.2015.03.028
Fanfrlík J, Bronowska AK, Řezáč J et al (2010) A reliable docking/scoring scheme based on the semiempirical quantum mechanical PM6-DH2 method accurately covering dispersion and H-bonding: HIV-1 protease with 22 ligands. J Phys Chem B 114:12666–12678. https://doi.org/10.1021/jp1032965
Wang S, Milne GWA, Nicklaus MC et al (1994) Protein kinase C. Modeling of the binding site and prediction of binding constants. J Med Chem 37:1326–1338. https://doi.org/10.1021/jm00035a013
Murray CW, Verdonk ML (2002) The consequences of translational and rotational entropy lost by small molecules on binding to proteins. J Comput Aided Mol Des 16:741–753. https://doi.org/10.1023/a:1022446720849
Stewart JJP (1990) Reviews in computational chemistry. Wiley, Hoboken
SantAnna CMR (2009) Molecular modeling methods in the study and design of bioactive compounds: an introduction. Rev Virtual Química. https://doi.org/10.5935/1984-6835.20090007
Menikarachchi L, Gascon J (2010) QM/MM approaches in medicinal chemistry research. Curr Top Med Chem 10:46–54. https://doi.org/10.2174/156802610790232297
Wollacott AM, Merz KM (2007) Assessment of semiempirical quantum mechanical methods for the evaluation of protein structures. J Chem Theory Comput 3:1609–1619. https://doi.org/10.1021/ct600325q
González R, Suárez CF, Bohórquez HJ et al (2017) Semi-empirical quantum evaluation of peptide: MHC class II binding. Chem Phys Lett 668:29–34. https://doi.org/10.1016/j.cplett.2016.12.015
Cabeza de Vaca I, Qian Y, Vilseck JZ et al (2018) Enhanced Monte Carlo methods for modeling proteins including computation of absolute free energies of binding. J Chem Theory Comput 14:3279–3288. https://doi.org/10.1021/acs.jctc.8b00031
Oliveira FG, Sant’Anna CMR, Caffarena ER et al (2006) Molecular docking study and development of an empirical binding free energy model for phosphodiesterase 4 inhibitors. Bioorg Med Chem 14:6001–6011. https://doi.org/10.1016/j.bmc.2006.05.017
LaMarche MJ, Borawski J, Bose A et al (2012) Anti-hepatitis C virus activity and toxicity of type III phosphatidylinositol-4-kinase beta inhibitors. Antimicrob Agents Chemother 56:5149–5156. https://doi.org/10.1128/AAC.00946-12
Waring MJ, Andrews DM, Faulder PF et al (2014) Potent, selective small molecule inhibitors of type III phosphatidylinositol-4-kinase α- but not β-inhibit the phosphatidylinositol signaling cascade and cancer cell proliferation. Chem Commun 50:5388–5390. https://doi.org/10.1039/C3CC48391F
Mejdrová I, Chalupská D, Kögler M et al (2015) Highly selective phosphatidylinositol 4-kinase IIIβ inhibitors and structural insight into their mode of action. J Med Chem 58:3767–3793. https://doi.org/10.1021/acs.jmedchem.5b00499
Mejdrová I, Chalupská D, Plačková P et al (2017) Rational design of novel highly potent and selective phosphatidylinositol 4-kinase IIIβ (PI4KB) inhibitors as broad-spectrum antiviral agents and tools for chemical biology. J Med Chem 60:100–118. https://doi.org/10.1021/acs.jmedchem.6b01465
Rutaganira FU, Fowler ML, McPhail JA et al (2016) Design and structural characterization of potent and selective inhibitors of phosphatidylinositol 4 kinase IIIβ. J Med Chem 59:1830–1839. https://doi.org/10.1021/acs.jmedchem.5b01311
Šála M, Kögler M, Plačková P et al (2016) Purine analogs as phosphatidylinositol 4-kinase IIIβ inhibitors. Bioorg Med Chem Lett 26:2706–2712. https://doi.org/10.1016/j.bmcl.2016.04.002
Halgren TA (1996) Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J Comput Chem 17:490–519
Stewart JJP (2007) Optimization of parameters for semiempirical methods V: modification of NDDO approximations and application to 70 elements. J Mol Model 13:1173–1213. https://doi.org/10.1007/s00894-007-0233-4
Berman HM (2000) The protein data bank. Nucleic Acids Res 28:235–242. https://doi.org/10.1093/nar/28.1.235
Burke JE, Inglis AJ, Perisic O et al (2014) Structures of PI4KIII complexes show simultaneous recruitment of Rab11 and its effectors. Science 344:1035–1038. https://doi.org/10.1126/science.1253397
Korb O, Stützle T, Exner TE (2009) Empirical scoring functions for advanced protein−ligand docking with PLANTS. J Chem Inf Model 49:84–96. https://doi.org/10.1021/ci800298z
Stewart JJP (2013) Optimization of parameters for semiempirical methods VI: more modifications to the NDDO approximations and re-optimization of parameters. J Mol Model 19:1–32. https://doi.org/10.1007/s00894-012-1667-x
Chambers CC, Hawkins GD, Cramer CJ, Truhlar DG (1996) Model for aqueous solvation based on class IV atomic charges and first solvation shell effects. J Phys Chem 100:16385–16398. https://doi.org/10.1021/jp9610776
Cer RZ, Mudunuri U, Stephens R, Lebeda FJ (2009) IC50-to-Ki: a web-based tool for converting IC50 to Ki values for inhibitors of enzyme activity and ligand binding. Nucleic Acids Res 37:W441–W445. https://doi.org/10.1093/nar/gkp253
Nunes IKC (2013) Novos inibidores de fosfodiesterases 4 análogos de LASSBio-488 – desenho, síntese e análise comparativa de suas propriedades físico-químicas e cinéticas. PhD Thesis, Universidade Federal do Rio de Janeiro
Lima L, Barreiro E (2005) Bioisosterism: a useful strategy for molecular modification and drug design. Curr Med Chem 12:23–49. https://doi.org/10.2174/0929867053363540
Lima LM, Barreiro EJ (2017) Beyond bioisosterism: new concepts in drug discovery. Comprehensive medicinal chemistry III. Elsevier, New York, pp 186–210
Gilson MK, Zhou H-X (2007) Calculation of protein-ligand binding affinities. Annu Rev Biophys Biomol Struct 36:21–42. https://doi.org/10.1146/annurev.biophys.36.040306.132550
Kukic P, Farrell D, McIntosh LP et al (2013) Protein dielectric constants determined from NMR chemical shift perturbations. J Am Chem Soc 135:16968–16976. https://doi.org/10.1021/ja406995j
Knight ZA, Feldman ME, Balla A et al (2007) A membrane capture assay for lipid kinase activity. Nat Protoc 2:2459–2466. https://doi.org/10.1038/nprot.2007.361
Keaney EP, Connolly M, Dobler M et al (2014) 2-Alkyloxazoles as potent and selective PI4KIIIβ inhibitors demonstrating inhibition of HCV replication. Bioorg Med Chem Lett 24:3714–3718. https://doi.org/10.1016/j.bmcl.2014.07.015
Lima LM, Frattani FS, dos Santos JL et al (2008) Synthesis and anti-platelet activity of novel arylsulfonate–acylhydrazone derivatives, designed as antithrombotic candidates. Eur J Med Chem 43:348–356. https://doi.org/10.1016/j.ejmech.2007.03.032
Silva TF (2012) Planejamento, síntese e avaliação farmacológica de uma nova série de derivados cicloalquil-N-Acilidrazonas análogos de LASSBio-294. MSc. Dissertation, Universidade Federal do Rio de Janeiro
Barbosa MLDC, Lima LM, Tesch R et al (2014) Novel 2-chloro-4-anilino-quinazoline derivatives as EGFR and VEGFR-2 dual inhibitors. Eur J Med Chem 71:1–14. https://doi.org/10.1016/j.ejmech.2013.10.058
Thota S, Rodrigues DA, de Pinheiro P et al (2018) N-Acylhydrazones as drugs. Bioorg Med Chem Lett 28:2797–2806. https://doi.org/10.1016/j.bmcl.2018.07.015
Thorne N, Auld DS, Inglese J (2010) Apparent activity in high-throughput screening: origins of compound-dependent assay interference. Curr Opin Chem Biol 14:315–324. https://doi.org/10.1016/j.cbpa.2010.03.020
Acknowledgements
The authors would like to thank the Brazilian funding agencies for the financial support involved in this work: Conselho Nacional de Desenvolvimento Científico e Tecnológico—CNPq (INCT-INOFAR grant #465.249/2014-0; grant #309229/2018-9); Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES)—Finance Code 001; Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro—FAPERJ (Grant E-26/202.146/2015); Additionally, we would like to thank the Biomedical Sciences Institute of the Rio de Janeiro Federal University (ICB-UFRJ) for the infrastructure provided to develop this work.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by NMC. The first draft of the manuscript was written by NMC and all authors revised and prepared following versions of the manuscript. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Colodette, N.M., Franco, L.S., Maia, R.C. et al. Novel phosphatidylinositol 4-kinases III beta (PI4KIIIβ) inhibitors discovered by virtual screening using free energy models. J Comput Aided Mol Des 34, 1091–1103 (2020). https://doi.org/10.1007/s10822-020-00327-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-020-00327-9