Abstract
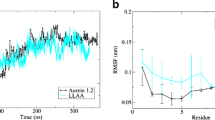
Human cathelicidin LL-37 has recently attracted interest as a potential therapeutic agent, mostly because of its ability to kill a wide variety of pathogens and cancer cells. In this study, we used molecular dynamics simulation aimed to get insights that help to correlate with the antibacterial activity of previously designed LL-37 anticancer derivative (i.e. GF-17). Two independent molecular dynamics simulation involving four units of GF-17 peptide in the mixture (9:1) of 1,2-dipalmitoyl-sn-glycero-3-phosphorylethanolamine (DPPE) and 1,2-dipalmitoyl-sn-glycero-3-phosphoglycerol (DPPG), and the pure DPPG lipids were performed. Various properties of membranes such as mass density distributions, area per lipid, bilayer thickness, and lateral diffusion were examined in both systems. The results showed that the thickness of the bilayer was not affected by the presence of GF-17, while the area per lipid and lateral diffusion of lipids showed an increase. Moreover, the potential of the mean force (PMF) method was used to calculate the free energy profile for transferring GF-17 from the bulk water into both kinds of membranes. It revealed that penetration of GF-17 into the DPPG membrane was more favorable than the DPPE/DPPG membrane, and there was no energy barrier for crossing through the bilayer center. Investigation of the radius of gyration (Rg) and root mean square fluctuation (RMSF) of peptides in two membranes showed that GF-17 had more compactness and rigidity in the pure DPPG system. By examining the secondary structure of GF-17 peptide, it was seen that the α-helix, and coil structures in both DPPE/DPPG and pure DPPG membranes are dominant.










Similar content being viewed by others
References
Guilhelmelli F, Vilela N, Albuquerque P, Derengowski L, Silva-Pereira I, Kyaw C (2013) Antibiotic development challenges: the various mechanisms of action of antimicrobial peptides and of bacterial resistance. Front Microbiol 4:353
Hale JDF, Hancock REW (2007) Alternative mechanisms of action of cationic antimicrobial peptides on bacteria. Expert Rev Anti Infect Ther 5:951–959
Memariani H, Shahbazzadeh D, Sabatier J-M, Memariani M, Karbalaeimahdi A, Bagheri KP (2016) Mechanism of action and in vitro activity of short hybrid antimicrobial peptide PV3 against Pseudomonas aeruginosa. Biochem Biophys Res Commun 479:103–108
Pachón-Ibáñez ME, Smani Y, Pachón J (2017) Perspectives for clinical use of engineered human host defense antimicrobial peptides. FEMS Microbiol Rev 41:323–342
Memariani H, Shahbazzadeh D, Sabatier J, Pooshang K, Bagheri (2018) Membrane-active peptide PV 3 efficiently eradicates multidrug‐resistant Pseudomonas aeruginosa in a mouse model of burn infection. Apmis 126:114–122
Akbari R, Vala MH, Hashemi A, Aghazadeh H, Sabatier J-M, Bagheri KP (2018) Action mechanism of melittin-derived antimicrobial peptides, MDP1 and MDP2, de novo designed against multidrug resistant bacteria. Amino Acids 50:1231–1243
Memariani H, Shahbazzadeh D, Ranjbar R, Behdani M, Memariani M et al (2017) Design and characterization of short hybrid antimicrobial peptides from pEM-2, mastoparan‐VT 1, and mastoparan‐B. Chem Biol Drug Des 89:327–338
Pashaei F, Bevalian P, Akbari R, Bagheri KP (2019) Single dose eradication of extensively drug resistant Acinetobacter spp. In a mouse model of burn infection by melittin antimicrobial peptide. Microb Pathog 127:60–69
Aghazadeh H, Memariani H, Ranjbar R, Pooshang K, Bagheri (2019) The activity and action mechanism of novel short selective LL-37‐derived anticancer peptides against clinical isolates of Escherichia coli. Chem Biol Drug Des 93:75–83
Lohner K (2009) New strategies for novel antibiotics: peptides targeting bacterial cell. Gen Physiol Biophys 28:105–116
Gaspar D, Veiga AS, Castanho MARB (2013) From antimicrobial to anticancer peptides. A review. Front Microbiol 4:294
Mahmoodzadeh A, Zarrinnahad H, Bagheri KP, Moradia A, Shahbazzadeh D (2015) First report on the isolation of melittin from Iranian honey bee venom and evaluation of its toxicity on gastric cancer AGS cells. J Chin Med Assoc 78:574–583
Zarrinnahad H, Mahmoodzadeh A, Hamidi MP, Mahdavi M, Moradi A, Bagheri KP, Shahbazzadeh D (2018) Apoptotic effect of melittin purified from Iranian honey bee venom on human cervical cancer HeLa cell line. Int J Pept Res Ther 24:563–570
Harris F, Dennison SR, Singh J, Phoenix DA (2013) On the selectivity and efficacy of defense peptides with respect to cancer cells. Med Res Rev 33:190–234
Wang L, Dong C, Li X, Han W, Su X (2017) Anticancer potential of bioactive peptides from animal sources. Oncol Rep 38:637–651
Hoskin DW, Ramamoorthy A (2008) Studies on anticancer activities of antimicrobial peptides. Biochim Biophys Acta 1778:357–375
van Zoggel H, Carpentier G, Dos Santos C, Hamma-Kourbali Y, Courty J, Amiche M, Delbé J (2012) Antitumor and angiostatic activities of the antimicrobial peptide dermaseptin B2. PLoS ONE 7:e44351
Sørensen OE, Follin P, Johnsen AH, Calafat J, Tjabringa GS, Hiemstra PS, Borregaard N (2001) Human cathelicidin, hCAP-18, is processed to the antimicrobial peptide LL-37 by extracellular cleavage with proteinase 3. Blood 97:3951–3959
Morizane S, Gallo RL (2012) Antimicrobial peptides in the pathogenesis of psoriasis. J Dermatol 39:225–230
Bandurska K, Berdowska A, Barczyńska-Felusiak R, Krupa P (2015) Unique features of human cathelicidin LL‐37. Biofactors 41:289–300
Tsai P-W, Cheng Y-L, Hsieh W-P, Lan C-Y (2014) Responses of Candida albicans to the human antimicrobial peptide LL-37. J Microbiol 52:581–589
Vandamme D, Landuyt B, Luyten W, Schoofs L (2012) A comprehensive summary of LL-37, the factotum human cathelicidin peptide. Cell Immunol 280:22–35
Jacob B, Park I, Bang J, Shin SY (2013) Short KR-12 analogs designed from human cathelicidin LL‐37 possessing both antimicrobial and antiendotoxic activities without mammalian cell toxicity. J Pept Sci 19:700–707
Kamysz E, Sikorska E, Jaśkiewicz M, Bauer M, Neubauer D, Bartoszewska S, Barańska-Rybak W, Kamysz W (2020) Lipidated analogs of the LL-37-derived peptide fragment KR12—structural analysis, surface-active properties and antimicrobial activity. Int J Mol Sci 21:887
Wang G, Watson KM, Buckheit RW (2008) Anti-human immunodeficiency virus type 1 activities of antimicrobial peptides derived from human and bovine cathelicidins. Antimicrob Agents Chemother 52:3438–3440
Noore J, Noore A, Li B (2013) Cationic antimicrobial peptide LL-37 is effective against both extra-and intracellular Staphylococcus aureus. Antimicrob Agents Chemother 57:1283–1290
Wang G, Mishra B, Epand RF, Epand RM (2014) High-quality 3D structures shine light on antibacterial, anti-biofilm and antiviral activities of human cathelicidin LL-37 and its fragments. Biochim Biophys Acta 1838:2160–2172
Wang X, Junior JCB, Mishra B, Lushnikova T, Epand RM, Wang G (2017) Arginine-lysine positional swap of the LL-37 peptides reveals evolutional advantages of the native sequence and leads to bacterial probe. Biochim Biophys Acta 1859:1350–1361
Mishra B, Golla RM, Lau K, Lushnikova T, Wang G (2015) Anti-staphylococcal biofilm effects of human cathelicidin peptides. ACS Med Chem Lett 7:117–121
Li X, Li Y, Han H, Miller DW, Wang G (2006) Solution structures of human LL-37 fragments and NMR-based identification of a minimal membrane-targeting antimicrobial and anticancer region. J Am Chem Soc 128:5776–5785
Orsi M, Essex JW (2010) Permeability of drugs and hormones through a lipid bilayer: insights from dual-resolution molecular dynamics. Soft Matter 6:3797–3808
Zhao L, Cao Z, Bian Y, Hu G, Wang J, Zhou Y (2018) Molecular dynamics simulations of human antimicrobial peptide LL-37 in model POPC and POPG lipid bilayers. Int J Mol Sci 19:1186
Mojumdar EH, Lyubartsev AP (2010) Molecular dynamics simulations of local anesthetic articaine in a lipid bilayer. Biophys Chem 153:27–35
Ganjali Koli M, Azizi K (2017) The partition and transport behavior of cytotoxic ionic liquids (ILs) through the DPPC bilayer: insights from molecular dynamics simulation. Mol Membr Biol 33:64–75. https://doi.org/10.1080/09687688.2017.1384859
Högberg C-J, Lyubartsev AP (2008) Effect of local anesthetic lidocaine on electrostatic properties of a lipid bilayer. Biophys J 94:525–531
Azizi K, Ganjali Koli M (2016) Molecular dynamics simulations of oxprenolol and propranolol in a DPPC lipid bilayer. J Mol Graph Model 64:153–164
Jahangiri S, Jafari M, Arjomand M, Mehrnejad F (2018) Molecular insights into the interactions of GF-17 with the gram‐negative and gram‐positive bacterial lipid bilayers. J Cell Biochem 119:9205–9216
Wang X, Mishra B, Lushnikova T, Narayana JL, Wang G (2018) Amino acid composition determines peptide activity spectrum and hot-spot‐based design of merecidin. Adv Biosyst 2:1700259
Rowlett VW, Mallampalli VKPS, Karlstaedt A, Dowhan W, Taegtmeyer H, Margolin W, Vitrac H (2017) The impact of membrane phospholipid alterations in Escherichia coli on cellular function and bacterial stress adaptation. J Bacteriol 199(13):e00849-16
Sohlenkamp C, Geiger O (2016) Bacterial membrane lipids: diversity in structures and pathways. FEMS Microbiol Rev 40:133–159
Malanovic N, Lohner K (2016) Gram-positive bacterial cell envelopes: the impact on the activity of antimicrobial peptides. Biochim Biophys Acta 1858:936–946
Jo S, Lim JB, Klauda JB, Im W (2009) CHARMM-GUI membrane builder for mixed bilayers and its application to yeast membranes. Biophys J 97:50–58
Lee J, Cheng X, Swails JM, Yeom MS, Eastman PK, Lemkul JA, Wei S, Buckner J, Jeong JC, Qi Y (2015) CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J Chem Theory Comput 12:405–413
Wu EL, Cheng X, Jo S, Rui H, Song KC, Dávila-Contreras EM, Qi Y, Lee J, Monje‐Galvan V, Venable RM (2014) CHARMM‐GUI membrane builder toward realistic biological membrane simulations. J Comput Chem 35:1997–2004
Berendsen HJC, van der Spoel D, van Drunen R (1995) GROMACS: a message-passing parallel molecular dynamics implementation. Comput Phys Commun 91:43–56
Pall S, Abraham MJ, Kutzner C, Hess B, Lindahl E (2014) Tackling exascale software challenges in molecular dynamics simulations with GROMACS. International conference exascale applied software. Springer, Berlin, pp 3–27
Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E (2015) GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1:19–25
Venable RM, Sodt AJ, Rogaski B, Rui H, Hatcher E, MacKerell AD Jr, Pastor RW, Klauda JB (2014) CHARMM all-atom additive force field for sphingomyelin: elucidation of hydrogen bonding and of positive curvature. Biophys J 107:134–145
Klauda JB, Venable RM, Freites JA, O’Connor JW, Tobias DJ, Mondragon-Ramirez C, Vorobyov I, MacKerell AD, Jr RW, Pastor (2010) Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J Phys Chem B 114:7830–7843
Boggara MB, Krishnamoorti R (2010) Partitioning of nonsteroidal antiinflammatory drugs in lipid membranes: a molecular dynamics simulation study. Biophys J 98:586–595
Parrinello M, Rahman A (1980) Crystal structure and pair potentials: a molecular-dynamics study. Phys Rev Lett 45:1196
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935
Hess B, Bekker H, Berendsen HJC, Fraaije JGEM (1997) LINCS: a linear constraint solver for molecular simulations. J Comput Chem 18:1463–1472
Hockney RW, Goel SP, Eastwood JW (1974) Quiet high-resolution computer models of a plasma. J Comput Phys 14:148–158
Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG (1995) A smooth particle mesh Ewald method. J Chem Phys 103:8577–8593
Snyman J (2005) Practical mathematical optimization: an introduction to basic optimization theory and classical and new gradient-based algorithms. Springer, Berlin
Torrie GM, Valleau JP (1977) Nonphysical sampling distributions in Monte Carlo free-energy estimation: umbrella sampling. J Comput Phys 23:187–199
Koli MG, Azizi K (2019) Investigation of benzodiazepines (BZDs) in a DPPC lipid bilayer: insights from molecular dynamics simulation and DFT calculations. J Mol Graph Model 90:171–179
Hub JS, De Groot BL, Van Der Spoel D (2010) A free weighted histogram analysis implementation including robust error and autocorrelation estimates. J Chem Theory Comput 6:3713–3720
Vivcharuk V, Kaznessis YN (2011) Thermodynamic analysis of protegrin-1 insertion and permeation through a lipid bilayer. J Phys Chem B 115:14704–14712
Epand RM, Vogel HJ (1999) Diversity of antimicrobial peptides and their mechanisms of action. Biochim Biophys Acta 1462:11–28
Matyus E, Kandt C, Tieleman DP (2007) Computer simulation of antimicrobial peptides. Curr Med Chem 14:2789–2798
Zhao W, Róg T, Gurtovenko AA, Vattulainen I, Karttunen M (2008) Role of phosphatidylglycerols in the stability of bacterial membranes. Biochimie 90:930–938
Bechinger B (2004) Structure and function of membrane-lytic peptides. CRC Crit Rev Plant Sci 23:271–292
Sato H, Feix JB (2006) Peptide–membrane interactions and mechanisms of membrane destruction by amphipathic α-helical antimicrobial peptides. Biochim Biophys Acta 1758:1245–1256
Wimley WC (2010) Describing the mechanism of antimicrobial peptide action with the interfacial activity model. ACS Chem Biol 5:905–917
Kolusheva S, Shahal T, Jelinek R (2000) Peptide–membrane interactions studied by a new phospholipid/polydiacetylene colorimetric vesicle assay. Biochemistry 39:15851–15859
Leontiadou H, Mark AE, Marrink SJ (2006) Antimicrobial peptides in action. J Am Chem Soc 128:12156–12161
Pan J, Heberle FA, Tristram-Nagle S, Szymanski M, Koepfinger M, Katsaras J, Kučerka N (1818) Molecular structures of fluid phase phosphatidylglycerol bilayers as determined by small angle neutron and X-ray scattering. Biochim Biophys Acta 2012:2135–2148
Kabsch W, Sander C (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolym Orig Res Biomol 22:2577–2637
Malkov S, Zivkovic MV, Beljanski MV, Zaric SD (2005) Correlations of amino acids with secondary structure types: connection with amino acid structure, ArXiv Prepr. q-Bio/0505046
Acknowledgement
The authors gratefully thank the use of School of Computer Science, Institute for Research in Fundamental Science (IPM) as the computations were done there.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
No potential conflict of interest was reported by the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Aghazadeh, H., Ganjali Koli, M., Ranjbar, R. et al. Interactions of GF-17 derived from LL-37 antimicrobial peptide with bacterial membranes: a molecular dynamics simulation study. J Comput Aided Mol Des 34, 1261–1273 (2020). https://doi.org/10.1007/s10822-020-00348-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-020-00348-4