Abstract
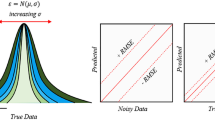
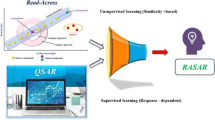
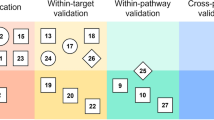
Notwithstanding the wide adoption of the OECD principles (or best practices) for QSAR modeling, disparities between in silico predictions and experimental results are frequent, suggesting that model predictions are often too optimistic. Of these OECD principles, the applicability domain (AD) estimation has been recognized in several reports in the literature to be one of the most challenging, implying that the actual reliability measures of model predictions are often unreliable. Applying tree-based error analysis workflows on 5 QSAR models reported in the literature and available in the QsarDB repository, i.e., androgen receptor bioactivity (agonists, antagonists, and binders, respectively) and membrane permeability (highest membrane permeability and the intrinsic permeability), we demonstrate that predictions erroneously tagged as reliable (AD prediction errors) overwhelmingly correspond to instances in subspaces (cohorts) with the highest prediction error rates, highlighting the inhomogeneity of the AD space. In this sense, we call for more stringent AD analysis guidelines which require the incorporation of model error analysis schemes, to provide critical insight on the reliability of underlying AD algorithms. Additionally, any selected AD method should be rigorously validated to demonstrate its suitability for the model space over which it is applied. These steps will ultimately contribute to more accurate estimations of the reliability of model predictions. Finally, error analysis may also be useful in “rational” model refinement in that data expansion efforts and model retraining are focused on cohorts with the highest error rates.



Similar content being viewed by others
Supporting availability of data and materials
Datasets and the python-based error analysis implementation employed in this work are available at: https://github.com/sjbarigye/erroranalysis. Original datasets, models and performance metrics may be accessed via the public QsarDB repository at : https://doi.org/10.15152/QDB.236, https://doi.org/10.15152/QDB.206.
References
Cherkasov A, Muratov EN, Fourches D, Varnek A, Baskin II, Cronin M, Dearden J, Gramatica P, Martin YC, Todeschini R (2014) J Med Chem 57(12):4977
Neves BJ, Braga RC, Melo-Filho CC, Moreira-Filho JT, Muratov EN, Andrade CH (2018) Front pharmacol 9
Sheridan RP (2013) J Chem Inf Model 53(4):783
Muratov EN, Bajorath J, Sheridan RP, Tetko IV, Filimonov D, Poroikov V, Oprea TI, Baskin II, Varnek A, Roitberg A (2020) Chem Soc Rev 49(11):3525
Tropsha A (2010) Mol Inf 29(6–7):476
Mathea M, Klingspohn W, Baumann K (2016) Mol Inf 35(5):160
Sahigara F, Mansouri K, Ballabio D, Mauri A, Consonni V, Todeschini R (2012) Molecules 17(5):4791
Tropsha A, Golbraikh A (2007) Curr Pharm Des 13(34):3494
Jaworska J, Nikolova-Jeliazkova N, Aldenberg T (2005) ATLA Altern Lab Anim 33(5):445
Tetko IV, Sushko I, Pandey AK, Zhu H, Tropsha A, Papa E, Oberg T, Todeschini R, Fourches D, Varnek A (2008) J Chem Inf Model 48(9):1733
Sheridan RP (2012) J Chem Inf Model 52(3):814
Sheridan RP (2013) J Chem Inf Model 53(11):2837
Norinder U, Carlsson L, Boyer S, Eklund M (2014) J Chem Inf Model 54(6):1596
Bosc N, Atkinson F, Felix E, Gaulton A, Hersey A, Leach AR (2019) J Cheminformatics 11:1
Cortes-Ciriano I, Murrell DS, van Westen GJ, Bender A, Malliavin TE (2015) J Cheminformatics 7(1):1
Oršolić D, Šmuc T (2023) Bioinformatics 39(8):btad465
Ruusmann V, Sild S, Maran U (2015) J Cheminformatics 7(1):32
Oja M, Sild S, Maran U (2019) J Chem Inf Model 59(5):2442
Piir G, Sild S, Maran U (2021) Chemosphere 262:128313
Wolpert DH, Macready WG (1997) IEEE T Evolut Comput 1(1):67
Sullivan K, Manuppello J, Willett C (2014) SAR QSAR Environ Res 25(5):357
Dearden JC, Rowe PH (2015) Use of artificial neural networks in the QSAR prediction of physicochemical properties and toxicities for REACH legislation. In: Cartwright H (ed) Artificial neural networks. Methods in Molecular Biology. Springer, New York, NY, p 65
Pavan M, Worth A (2008) SAR QSAR Environ Res 19(7–8):785
Miller TH, Gallidabino MD, MacRae JI, Hogstrand C, Bury NR, Barron LP, Snape JR, Owen SF (2018) Environ Sci Technol 52(22):12953
Gouin T (2010) Environ Sci Policy 13(3):175
Syberg K, Hansen SF (2016) Sci Total Environ 541:784
Scior T, Medina-Franco J, Do Q-T, Martínez-Mayorga K, Yunes Rojas J, Bernard P (2009) Curr Med Chem 16(32):4297
Martin YC (2012) Wiley Interdisciplinary Reviews. Comput Mol Sci 2(3):435
Gini G (2018) QSAR: what else? Computational toxicology: methods and protocols, vol 1800. Humana, New York, NY, p 79
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
J.R.M: investigation, formal analysis, writing – original draft, visualization. E.A.M: methodology, validation, formal analysis. N.P.P: resources, conceptualization. E.C.T: software implementation & scripting, Y.P.C: investigation, data curation, methodology. G.A.C: software implementation, writing – review & editing, F.M.R: data curation, formal analysis. Y.M.P: methodology, writing – review & editing, project administration. S.J.B: conceptualization, formal analysis, methodology, writing - review & editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mora, J.R., Marquez, E.A., Pérez-Pérez, N. et al. Rethinking the applicability domain analysis in QSAR models. J Comput Aided Mol Des 38, 9 (2024). https://doi.org/10.1007/s10822-024-00550-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10822-024-00550-8