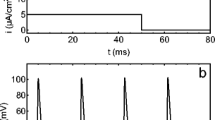
Abstract
We present an event tree analysis of studying the dynamics of the Hodgkin-Huxley (HH) neuronal networks. Our study relies on a coarse-grained projection to event trees and to the event chains that comprise these trees by using a statistical collection of spatial-temporal sequences of relevant physiological observables (such as sequences of spiking multiple neurons). This projection can retain information about network dynamics that covers multiple features, swiftly and robustly. We demonstrate that for even small differences in inputs, some dynamical regimes of HH networks contain sufficiently higher order statistics as reflected in event chains within the event tree analysis. Therefore, this analysis is effective in discriminating small differences in inputs. Moreover, we use event trees to analyze the results computed from an efficient library-based numerical method proposed in our previous work, where a pre-computed high resolution data library of typical neuronal trajectories during the interval of an action potential (spike) allows us to avoid resolving the spikes in detail. In this way, we can evolve the HH networks using time steps one order of magnitude larger than the typical time steps used for resolving the trajectories without the library, while achieving comparable statistical accuracy in terms of average firing rate and power spectra of voltage traces. Our numerical simulation results show that the library method is efficient in the sense that the results generated by using this numerical method with much larger time steps contain sufficiently high order statistical structure of firing events that are similar to the ones obtained using a regular HH solver. We use our event tree analysis to demonstrate these statistical similarities.













Similar content being viewed by others
References
Abraham, N. M., et al. (2004). Maintaining accuracy at the expense of speed: Stimulus similarity defines odor discrimination time in mice. Neuron, 44, 865–876.
Cai, D., Rangan, A. V., & McLaughlin, D. W. (2005). Architectural and synaptic mechanisms underlying coherent spontaneous activity in v1. Proceedings of the National Academy of Sciences of the United States of America, 102, 5868–5873.
Dayan, P., & Abbott, L. F. (2001). Theoretical neuroscience: Computational and mathematical modeling of neural systems. Cambridge, MA: MIT Press.
Gardiner, C. W. (1998). A handbook of stochastic methods. Berlin: Springer.
Gear, C. W. (1971). Numerical initial value problems in ordinary differential equations. Englewood Cliffs, NJ: Prentice Hall.
Hansel, D., Mato, G., Meunier, C., & Neltner, L. (1998) On numerical simulations of integrate-and-fire neural networks. Neural Computation, 10, 467–483.
Hodgkin, A. L., & Huxley, A. F. (1952). A quantitative description of membrane current and its application to conduction and excitation in nerve. Journal of Physiology, 117, 500–544.
Litvak, V., Sompolinsky, H., Segev, I., & Abeles, M. (2003). On the transmission of rate code in long feedforward networks with excitatory–inhibitory balance. Journal of Neuroscience, 23, 3006–3015.
Mainen, Z. F. (2006). Behavioral analysis of olfactory coding and computation in rodents. Current Opinion in Neurobiology, 16, 429–434.
Mattia, M., & Del Giudice, P. (2000). Efficient event-driven simulation of large networks of spiking neurons and dynamical synapses. Neural Computation, 12, 2305–2329.
McLaughlin, D., Shapley, R., Shelley, M., & Wielaard, J. (2000). A neuronal network model of macaque primary visual cortex (V1): Orientation selectivity and dynamics in the input layer 4Cα. Proceedings of the National Academy of Sciences of the United States of America, 97, 8087–8092.
Rangan, A. V., & Cai, D. (2007). Fast numerical methods for simulating large-scale integrate-and-fire neuronal networks. Journal of Computational Neuroscience, 22, 81–100.
Rangan, A. V., Cai, D., & McLaughlin D. W. (2005). Modeling the spatiotemporal cortical activity associated with the line-motion illusion in primary visual cortex. Proceedings of the National Academy of Sciences of the United States of America, 102, 18793–18800.
Rangan, A. V., Cai, D., & McLaughlin, D. W. (2008). Quantifying neuronal network dynamics through coarse-grained event trees. Proceedings of the National Academy of Sciences of the United States of America, 105, 10990–10995.
Reutimann, J., Giugliano, M., & Fusi, S. (2003). Event-based simulation of spiking neurons with stochastic dynamics. Neural Computation, 15, 811–830.
Roitman, J. D., & Shadlen, M. N. (2002). Response of neurons in the lateral intraparietal area during a combined visual discrimination reaction time task. Journal of Neuroscience, 22, 9475–9489.
Rousselet, G. A., Mace, M. J., & Fabre-Thorpe, M. (2003). Is it an animal? Is it a human face? Fast processing in upright and inverted natural scenes. Journal of Visualization, 3, 440–455.
Rudolph, M., & Destexhe, A. (2007). How much can we trust neural simulation strategies? Neurocomputing, 70, 1966–1969.
Schuster, H. G., & Just, W. (2005). Deterministic Chaos. Weinheim: Wiley-VCH Verlag.
Shadlen, M. N., & Newsome, W. T. (1998). The variable discharge of cortical neurons: Implications for connectivity, computation, and information coding. Journal of Neuroscience, 18, 3870–3896.
Shelley, M. J., & Tao, L. (2001). Efficient and accurate time-stepping schemes for integrate-and-fire neuronal networks. Journal of Computational Neuroscience, 11, 111–119.
Somers, D., Nelson, S., & Sur, M. (1995). An emergent model of orientation selectivity in cat visual cortical simple cells. Journal of Neuroscience, 15, 5448–5465.
Sun, Y., Zhou, D., Rangan, A. V., & Cai, D. (2009). Library-based numerical reduction of the Hodgkin-Huxley neuron for network simulation. Journal of Computational Neuroscience, 27, 369–390.
Sun, Y., Zhou, D., Rangan, A. V., & Cai, D. (2010). Pseudo-Lyapunov exponents and predictability of HH neuronal network dynamics. Journal of Computational Neuroscience, 28, 247–266.
Thorpe, S. J., & Gautrais, J. (1998). Rank order coding. In: J. Bower (Ed.), Computational neuroscience: Trends in research (pp. 113–119). New York: Plenum.
Troyer, T., Krukowski, A., Priebe, N., & Miller, K. (1998). Contrast invariant orientation tuning in cat visual cortex with feedforward tuning and correlation based intracortical connectivity. Journal of Neuroscience, 18, 5908–5927.
Uchida, N., & Mainen, Z. F. (2003). Speed and accuracy of olfactory discrimination in the rat. Nature Neuroscience, 6, 1224–1229.
Uchida, N., Kepecs, A., & Mainen, Z. F. (2006). Seeing at a glance, smelling in a whiff: Rapid forms of perceptual decision making. Nature Reviews. Neuroscience, 7, 485–491.
Victor, J. D., & Purpura, K. P. (1997). Sensory coding in cortical neurons: Recent results and speculations. Annals of the New York Academy of Sciences, 835, 330–352.
Acknowledgements
The work was supported by NSF grants DMS-0506396, DMS-0507901, DMS-1009575 and a grant from the Swartz foundation. Y. Sun was supported by NSF Joint Institutes’ Postdoctoral Fellowship through SAMSI. D. Zhou was supported by Shanghai Pujiang Program (Grant No. 10PJ1406300) and NSFC (Grant No. 11026052). We thank two anonymous referees for helpful comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Action Editor: David Terman
Appendices
Appendix A: Parameter values for the Hodgkin-Huxley equations
Parameter values or ranges and function definitions of the Hodgkin-Huxley model are as follows (Dayan and Abbott 2001):
Appendix B: Numerical method for a single neuron
Here we provide details of the numerical method for evolving the dynamics of a single neuron. For simplicity, we use vector X i to represent all the variables in the solution of the ith neuron:
Given an initial time t 0 and time step Δt, initial values X i (t 0), and spike times \(T^{\rm F}_{i,k}\) and \(T^{\rm S}_{j\neq i,k}\) from the other neurons in the network, a preset threshold value V th at which an action potential starts, our method computes a numerical solution of all variables X i (t 0 + Δt) as well as the intervening spike times \(T^{\rm S}_{i,k}\) (if any occurred) for the ith neuron as follows:
Algorithm 1. (Single neuron scheme)
Step 1: Input: an initial time t 0 , a time step Δt, a set of spike times \(T^{\rm F}_{i,k}\) and \(T^{\rm S}_{j\neq i,k}\) and associated strengths \(F_i^{\rm Q}\) and \(\widetilde{S}_{i,j}^{\rm Q}\).
Step 2: Consider the time interval [t 0, t 0 + Δt]. Let M denote the total number of feedforward and presynaptic spikes within this interval. Sort these spikes into an increasing list of M spike times \(T^{\rm sorted}_m\) with corresponding spike strengths \(S^{\rm sorted, Q}_m\) . In addition, we extend this notation such that \(T^{\rm sorted}_0\) : = t 0 , \(T^{\rm sorted}_{M+1}:= t_0 + \Delta t\) and \(S^{\rm sorted, Q}_0 =S^{\rm sorted, Q}_{M+1}:= 0\).
Step 3: For m = 1, ..., M + 1, advance the equations for the HH neuron model and its conductances (Eqs. ( 1 )–( 7 )) from \(T^{\rm sorted}_{m-1}\) to \(T^{\rm sorted}_m\) using the standard RK4 scheme to obtain \(X_i(T^{\rm sorted}_m)\) ; Then, update the conductance \(\widetilde{G}_i^{\rm Q}(T^{\rm sorted}_m)\) by adding the appropriate strengths \(S^{\rm sorted, Q}_m\).
Step 4: If the calculated values for \(V_i(T^{\rm sorted}_m)\) are each less than V th , then we can accept \(X_i(T^{\rm sorted}_{M+1})\) as the solution X i (t 0 + Δt). We update t 0 ← t 0 + Δt and return to step 2 and continue.
Step 5: Otherwise, let \(V_i(T^{\rm sorted}_m)\) be the first calculated voltage greater than V th . We know that the ith neuron fired somewhere during the interval \([T^{\rm sorted}_{m-1}, T^{\rm sorted}_m]\).
Step 6: In this case we use a high-order polynomial interpolation to find an approximation of the spike time t fire in the interval \([T^{\rm sorted}_{m-1}, T^{\rm sorted}_m]\) . For example, we can use the numerical values of \(V_i(T^{\rm sorted}_{m-1})\) , \(V_i(T^{\rm sorted}_m)\) , \(\frac{d}{dt}V_i(T^{\rm sorted}_{m-1})\) , \(\frac{d}{dt}V_i(T^{\rm sorted}_m)\) to form a cubic polynomial. We record t fire as the (k + 1)th postsynaptic spike time \(T^{\rm S}_{i,k+1}\) of the ith neuron. We update t 0 ← t 0 + Δt and return to step 2 and continue.
Appendix C: The library-based numerical method
Here, we briefly outline Algorithm 2, which uses the library to recover the spike. Given an initial time t 0 and time step Δt, initial values X i (t 0), and spike times \(T^{\rm F}_{i,k}\) and \(T^{\rm S}_{j\neq i,k}\) from the other neurons in the network, our method calculates a numerical solution of all variables X i (t 0 + Δt) as well as the intervening spike times \(T^{\rm S}_{i,k}\) (if any occurred) for the ith neuron as follows.
In order to build the data library, we choose N I different values of I input equally distributed in its range from 7.5 to 50.0 μA/cm2, which can essentially cover all typical values of I input of the spiking neurons at the moments when they fire in our network simulations. We also equally distribute N lib points of m input, h input, n input in their parameter ranges, respectively. These ranges should cover all typical values of m, h, n of a neuron when it fires in the HH network.
Algorithm 2. (Library algorithm)
Step 0: Pre-compute the data library of V, m, h, n for N I different values of the constant input current I input using very fine time step δ t. For each I input , we isolate an action potential form that starts exactly from V th to a later time (after T ref ms) where the membrane potential drops down around its minimum. Then we use this action potential form (i.e., the intermediate replica of the membrane potential) with different values of m input, h input, n input as initial values to compute the intermediate replica for m , h , n individually. In particular, we can obtain the reset values m re , h re , n re at the end point where the form of the action potential terminates. For each case of I input , there are N lib data sets of m, h, n, respectively.
Step 1: Input: the library, an initial time t 0 , a large time step Δt, a set of spike times \(T^{\rm F}_{i,k}\) and \(T^{\rm S}_{j\neq i,k}\) and associated strengths \(F_i^{\rm Q}\) and \(\widetilde{S}_{i,j}^{\rm Q}\).
Step 2: Consider the time interval [t 0, t 0 + Δt]. Let M denote the total number of feedforward and presynaptic spikes within this interval. Sort these spikes into an increasing list of M spike times \(T^{\rm sorted}_m\) with corresponding spike strengths \(S^{\rm sorted, Q}_m\) . In addition, we extend this notation such that \(T^{\rm sorted}_0\) : = t 0 , \(T^{\rm sorted}_{M+1}:= t_0 + \Delta t\) and \(S^{\rm sorted, Q}_0 =S^{\rm sorted, Q}_{M+1}:= 0\).
Step 3: For m = 1, ..., M + 1, advance the equations for the HH neuron model and its conductances (Eqs. ( 1 )–( 7 )) from \(T^{\rm sorted}_{m-1}\) to \(T^{\rm sorted}_m\) using the standard RK4 scheme to obtain \(X_i(T^{\rm sorted}_m)\) ; Then, update the conductance \(\widetilde{G}_i^{\rm Q}(T^{\rm sorted}_m)\) by adding the appropriate strengths \(S^{\rm sorted, Q}_m\).
Step 4: If the calculated values for \(V_i(T^{\rm sorted}_m)\) are each less than V th , then we can accept \(X_i(T^{\rm sorted}_{M+1})\) as the solution X i (t 0 + Δt). We update t 0 ← t 0 + Δt and return to step 2 and continue.
Step 5: Otherwise, let \(V_i(T^{\rm sorted}_m)\) be the first calculated voltage greater than V th . We know that the ith neuron fired somewhere during the interval \([T^{\rm sorted}_{m-1}, T^{\rm sorted}_m]\).
Step 6: In this case we use a high-order polynomial interpolation to find an approximation to the spike time t fire in the interval \([T^{\rm sorted}_{m-1}, T^{\rm sorted}_m]\) . For example, we can use the numerical values of \(V_i(T^{\rm sorted}_{m-1})\) , \(V_i(T^{\rm sorted}_m)\) , \(\frac{d}{dt}V_i(T^{\rm sorted}_{m-1})\) , \(\frac{d}{dt}V_i(T^{\rm sorted}_m)\) to form a cubic polynomial. We record t fire as the (k + 1)th postsynaptic spike time \(T^{\rm S}_{i,k+1}\) of the ith neuron.
Step 7: We compute the values of \(I^{\rm th} = - \sum_{\rm Q} G_i^{\rm Q}(t^{\rm f\/ire})\times\) \((V^{\rm th} - V_{\rm G}^{\rm Q}),\) as well as the gating variables m th , h th and n th at this time. Then, we perform a linear interpolation to find the corresponding reset values of V re, m re, h re, n re in the library. Meanwhile, we stop evolving Eqs. ( 1 )–( 5 ) for the next T ref ms, but evolve Eqs. ( 6 ) and ( 7 ) for the conductance terms \(G_i^{\rm Q}(t)\) and \(\widetilde{G}_i^{\rm Q}(t)\) as usual. We update \(t_0\leftarrow \min(t^{\rm f\/ire}+T^{\rm ref}\) , t + Δt) and return to step 2 and continue with the reset values V re, m re, h re, n re as the initial values V i (t 0), m i (t 0), h i (t 0) and n i (t 0).
Appendix D: More details on discriminability
Here we provide details of several definitions we used in Section 3.2. Although the occurrence count rate n takes only discrete values as shown in the x-axis of Fig. 6, it is more convenient to treat n as a continuous variable for the following discussion. Given a single sample T obs observation of the network dynamics, our discrimination task is to determine a possible candidate stimulus by using the two distributions P 1(n) and P 2(n) of an event chain. A simple procedure is to obtain the occurrence rate n of the event chain in the sample observation and compare it to a threshold number z. If n > z, we report I 1; otherwise we report I 2. Figure 6 shows that if we choose z to lie somewhere between the two distributions, say, the location where P 1(z) = P 2(z), this procedure can give the correct answer when the two distributions are fairly well separated, but will have difficulty discriminating the stimuli if their distributions merge together. This difficulty is clearly related to the degree to which the two distributions overlap.
First, we show how to define and calculate the hit rate A. The probability that the procedure will make the correct choice (called a hit) under the stimulus I 1 is the conditional probability that n > z given the stimulus I 1 was presented, \(\alpha:=P(n>z|I_1)=\int_z^\infty P_1(n)dn\). The probability that it will give the answer I 1 when the stimulus was actually I 2 (called a false alarm) is similarly \(\beta:=P(n>z|I_2)=\int_z^\infty P_2(n)dn\). These two probabilities completely determine the performance of the discrimination procedure because the probabilities for the other two cases (choosing I 2 when the correct answer is I 1, choosing I 2 when the correct answer is I 2) are \(1-\alpha=\int_0^z P_1(n)dn\) and \(1-\beta=\int_0^z P_2(n)dn\), respectively. Therefore, by applying this procedure to all different independent single samples in which the two stimuli I 1 and I 2 occur with equal probability, the overall probability of making a correct choice with the even chain \(\{\sigma^{j_1} \rightarrow \ldots \rightarrow \sigma^{j_m}\}\), i.e., the hit rate is given by
and the false alarm rate is
With A and B in hand, we measure the separation between the two distributions by using the information ratio, which is defined as \(I_{I_1,I_2}^{\sigma^{j_1} \rightarrow \ldots \rightarrow \sigma^{j_m}} \equiv A/B\). The information ratio is equal to one if the two distributions totally overlap (i.e., \(A=B=\frac{1}{2}\)), and it approaches to + ∞ when the two distributions are totally separated (i.e., A = 1, B = 0). Therefore, when we classify the stimulus by using the occurrence count of every event chain within an event tree, the “vote” of each contributing event chain for either stimulus I 1 or I 2 is weighted with the factor \(\log\left(I_{I_1,I_2}^{\sigma^{j_1} \rightarrow \ldots \rightarrow \sigma^{j_m}}\right)\). Then the weighted votes across the entire event tree are summed up to determine the candidate stimulus underlying the sample observation. This is similar to the concept of entropy in the information theory. For instance, if there are total N c event chains within an event tree and the vote of each chain \(\mathcal C_i\) is denoted by v i , which takes either value + 1 for stimulus I 1 or − 1 for I 2, the summed vote v s is given by
We choose I 1 if v s > 0, otherwise we choose I 2. If the two distributions of an event chain \(\mathcal C_i\) totally merge, its vote is void because its weight \(\log\left(I_{I_1,I_2}^{\mathcal C_i}\right) =0\). Thus, the final choice is made by the votes with relatively significant weights.
Finally, we apply the above procedure with the information of the entire tree to all different independent single samples. In a similar way as defining the hit rate above, the discriminability of the event tree is defined as the percentage of sample observations that were correctly classified under our voting procedure (choosing I 1 when the correct answer is I 1, choosing I 2 when the correct answer is I 2). The formula is given by D = N correct/N sample where N sample is the total number of sample observations and N correct is the number of times of making correct choices. In Figs. 10–13 in Section 4.2, we present the results by showing the discriminability as a function of m max.
Rights and permissions
About this article
Cite this article
Sun, Y., Rangan, A.V., Zhou, D. et al. Coarse-grained event tree analysis for quantifying Hodgkin-Huxley neuronal network dynamics. J Comput Neurosci 32, 55–72 (2012). https://doi.org/10.1007/s10827-011-0339-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10827-011-0339-7