Abstract
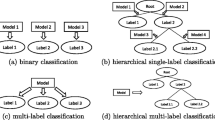
We address the task of hierarchical multi-label classification (HMC). HMC is a task of structured output prediction where the classes are organized into a hierarchy and an instance may belong to multiple classes. In many problems, such as gene function prediction or prediction of ecological community structure, classes inherently follow these constraints. The potential for application of HMC was recognized by many researchers and several such methods were proposed and demonstrated to achieve good predictive performances in the past. However, there is no clear understanding when is favorable to consider such relationships (hierarchical and multi-label) among classes, and when this presents unnecessary burden for classification methods. To this end, we perform a detailed comparative study over 8 datasets that have HMC properties. We investigate two important influences in HMC: the multiple labels per example and the information about the hierarchy. More specifically, we consider four machine learning tasks: multi-label classification, hierarchical multi-label classification, single-label classification and hierarchical single-label classification. To construct the predictive models, we use predictive clustering trees (a generalized form of decision trees), which are able to tackle each of the modelling tasks listed. Moreover, we investigate whether the influence of the hierarchy and the multiple labels carries over for ensemble models. For each of the tasks, we construct a single tree and two ensembles (random forest and bagging). The results reveal that the hierarchy and the multiple labels do help to obtain a better single tree model, while this is not preserved for the ensemble models.








Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Notes
Note that the hierarchical single-label classification models will be similar to the single-label classification models, with the difference that the predictive models are organized into a hierarchical architecture. This makes the interpretation of the HSC models an even more difficult task.
References
Alaydie, N., Reddy, C.K., Fotouhi, F. (2012). Exploiting label dependency for hierarchical multi-label classification. In Proceedings of the 16th Pacific-Asia conference on advances in knowledge discovery and data mining (pp. 294–305). Berlin: Heidelberg, New York: Springer.
Bakır, G.H., Hofmann, T., Schölkopf, B., Smola, A.J., Taskar, B., Vishwanathan, S.V.N. (Eds.) (2007). Predicting structured data. Cambridge, MA: The MIT Press.
Barros, R.C., Cerri, R., Freitas, A.A., de Carvalho, A.C.P.L.F. (2013). Probabilistic clustering for hierarchical multi-label classification of protein functions. In H. Blockeel, K. Kersting, S. Nijssen, F. železný (Eds.), Machine learning and knowledge discovery in databases, Lecture Notes in Computer Science, (Vol. 8189 pp. 385–400). Berlin Heidelberg: Springer.
Barutcuoglu, Z., Schapire, R.E., Troyanskaya, O.G. (2006). Hierarchical multi-label prediction of gene function. Bioinformatics, 22(7), 830–836.
Bauer, E., & Kohavi, R. (1999). An empirical comparison of voting classification algorithms: Bagging, boosting, and variants. Machine Learning, 36(1), 105–139.
Bi, W., & Kwok, J.T. (2012). Hierarchical multilabel classification with minimum bayes risk. In Proceedings of the 12th international conference on data mining (pp. 101–110).
Blockeel, H. (1998). Top-down induction of first order logical decision trees. Ph.D. thesis, Katholieke Universiteit Leuven, Leuven, Belgium.
Blockeel, H., Bruynooghe, M., Džeroski, S., Ramon, J., Struyf, J. (2002). Hierarchical multi-classification. In Proceedings of the ACM SIGKDD workshop on multi-relational data mining (pp. 21–35).
Blockeel, H., Schietgat, L., Struyf, J., Džeroski, S., Clare, A. (2006). Decision trees for hierarchical multilabel classification: A case study in functional genomics. In Knowledge discovery in databases: PKDD, Lecture Notes in Computer Science (Vol. 4213 pp. 18–29). Berlin Heidelberg: Springer.
Blockeel, H., & Struyf, J. (2002). Efficient algorithms for decision tree cross-validation. Journal of Machine Learning Research, 3, 621–650.
Breiman, L. (1996). Bagging predictors. Machine Learning, 24(2), 123–140.
Breiman, L. (2001). Random forests. Machine Learning, 45(1), 5–32.
Breiman, L., Friedman, J., Olshen, R.A., Stone, C.J. (1984). Classification and regression trees. London, UK: Chapman & Hall/CRC.
Cerri, R., Barros, R.C., de Carvalho, A.C.P.L.F. (2012). A genetic algorithm for hierarchical multi-label classification. In Proceedings of the 27th annual ACM symposium on applied computing (pp. 250–255).
Cerri, R., Barros, R.C., de Carvalho, A.C.P.L.F. (2014). Hierarchical multi-label classification using local neural networks. Journal of Computer and System Sciences, 80(1), 39–56.
Clare, A. (2003). Machine learning and data mining for yeast functional genomics. Ph.D. thesis, University of Wales Aberystwyth, Aberystwyth, UK.
Clare, A., & King, R.D. (2003). Predicting gene function in Saccharomyces cerevisiae. Bioinformatics, 19(S2), ii42–49.
Davis, J., & Goadrich, M. (2006). The relationship between precision-recall and ROC curves. In Proceedings of the 23rd international conference on machine learning (pp. 233–240).
Demšar, D., Džeroski, S., Larsen, T., Struyf, J., Axelsen, J., Bruns-Pedersen, M., Krogh, P.H. (2006). Using multi-objective classification to model communities of soil. Ecological Modelling, 191(1), 131–143.
Demšar, J. (2006). Statistical comparisons of classifiers over multiple data sets. Journal of Machine Learning Research, 7, 1–30.
Dietterich, T.G., Domingos, P., Getoor, L., Muggleton, S., Tadepalli, P. (2008). Structured machine learning: The next ten years. Machine Learning, 73(1), 3–23.
Dimitrovski, I., Kocev, D., Loskovska, S., Džeroski, S. (2008). Hierchical annotation of medical images. In Proceedings of the 11th international multiconference - information society (pp. 174–181). Ljubljana:JSI.
Džeroski, S. (2009). Machine learning applications in habitat suitability modeling. In: S.E. Haupt, A. Pasini, C. Marzban (Eds.) In Artificial intelligence methods in the environmental sciences. Springer Netherlands, (pp. 397–412).
Džeroski, S., Demšar, D., Grbović, J. (2000). Predicting chemical parameters of river water quality from bioindicator data. Applied Intelligence, 13(1), 7–17.
Estruch, V., Ferri, C., Hernández-Orallo, J., Ramírez-Quintana, M.J. (2006). Web categorisation using distance-based decision trees. Electronic Notes in Theoretical Computer Science, 157(2), 35–40.
Guan, Y., Myers, C.L., Hess, D.C., Barutcuoglu, Z., Caudy, A., Troyanskaya, O. (2008). Predicting gene function in a hierarchical context with an ensemble of classifiers. Genome Biology, 9(S1), S3+.
Kiritchenko, S., Famili, F., Matwin, S., Nock, R. (2006). Learning and evaluation in the presence of class hierarchies: Application to text categorization. In L. Lamontagne, M. Marchand (Eds.), Advances in artificial intelligence, Lecture Notes in Computer Science, (Vol. 4013 pp. 395–406). Berlin Heidelberg: Springer.
Klimt, B., & Yang, Y. (2004). The enron corpus: A new dataset for email classification research. In J.F. Boulicaut, F. Esposito, F. Giannotti, D. Pedreschi (Eds.), Machine learning: ECML, Lecture Notes in Computer Science, (Vol. 3201 pp. 217–226). Berlin Heidelberg: Springer.
Kocev, D., Vens, C., Struyf, J., Džeroski, S. (2013). Tree ensembles for predicting structured outputs. Pattern Recognition, 46(3), 817–833.
Kriegel, H.P., Borgwardt, K., Kröger, P., Pryakhin, A., Schubert, M., Zimek, A. (2007). Future trends in data mining. Data Mining and Knowledge Discovery, 15, 87–97.
Lehmann, T., Schubert, H., Keysers, D., Kohnen, M., Wein, B. (2003). The IRMA code for unique classification of medical images. In Medical imaging: PACS and integrated medical information systems: Design and evaluation (pp. 440–451).
Levatić, J., Kocev, D., Džeroski, S. (2013). The use of the label hierarchy in hmc improves performance: A case study in predicting community structure in ecology. In Proceedings of the workshop on new frontiers in mining complex patterns held in conjunction with ECML/PKDD2013 (pp. 189–201).
Levatić, J., Kocev, D., Džeroski, S. (2014). The use of the label hierarchy in hierarchical multi-label classification improves performance. In A. Appice, et al. (Eds.), New frontiers in mining complex patterns, Lecture Notes in Computer Science, (Vol. 8399 pp. 1–16): Springer International Publishing.
Lewis, D.D., Yang, Y., Rose, T.G., Li, F. (2004). RCV1: A new benchmark collection for text categorization research. Journal of Machine Learning Research, 5, 361–397.
Obozinski, G., Lanckriet, G., Grant, C., Jordan, M.I., Noble, W.S. (2008). Consistent probabilistic outputs for protein function prediction. Genome Biology, 9(S1), S6+.
Otero, F.E., Freitas, A.A., Johnson, C.G. (2010). A hierarchical multi-label classification ant colony algorithm for protein function prediction. Memetic Computing, 2(3), 165–181.
Quinlan, J.R. (1993). C4. 5: Programs for machine learning Vol. 1. San Francisco, CA: Morgan Kaufmann.
Rousu, J., Saunders, C., Szedmak, S., Shawe-Taylor, J. (2006). Kernel-based learning of hierarchical multilabel classification models. The Journal of Machine Learning Research, 7, 1601–1626.
Ruepp, A., Zollner, A., Maier, D., Albermann, K., Hani, J., Mokrejs, M., Tetko, I., Güldener, U., Mannhaupt, G., Münsterkötter, M., et al. (2004). The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Research, 32(18), 5539–5545.
Schietgat, L., Vens, C., Struyf, J., Blockeel, H., Kocev, D., Džeroski, S. (2010). Predicting gene function using hierarchical multi-label decision tree ensembles. BMC Bioinformatics, 11(2), 1–14.
Seni, G., & Elder, J.F. (2010). Ensemble methods in data mining: Improving accuracy through combining predictions: Morgan & Claypool Publishers.
Silla, C., & Freitas, A. (2011). A survey of hierarchical classification across different application domains. Data Mining and Knowledge Discovery, 22(1-2), 31–72.
Silla, C.N., & Freitas, A.A. (2009). A global-model naive bayes approach to the hierarchical prediction of protein functions. In Proceeding of the 9th IEEE international conference on data mining (pp. 992–997).
Slavkov, I., Gjorgjioski, V., Struyf, J., Džeroski, S. (2010). Finding explained groups of time-course gene expression profiles with predictive clustering trees. Molecular BioSystems, 6(4), 729–740.
Valentini, G. (2011). True path rule hierarchical ensembles for genome-wide gene function prediction. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 8(3), 832–847.
Vens, C., Struyf, J., Schietgat, L., Džeroski, S., Blockeel, H. (2008). Decision trees for hierarchical multi-label classification. Machine Learning, 73(2), 185–214.
Acknowledgments
We would like to acknowledge the support of the European Commission through the project MAESTRA - Learning from Massive, Incompletely annotated, and Structured Data (Grant number ICT-2013-612944).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Levatić, J., Kocev, D. & Džeroski, S. The importance of the label hierarchy in hierarchical multi-label classification. J Intell Inf Syst 45, 247–271 (2015). https://doi.org/10.1007/s10844-014-0347-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10844-014-0347-y