Abstract
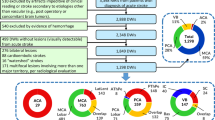
Statistical analysis of magnetic resonance angiography (MRA) brain artery trees is performed using two methods for mapping brain artery trees to points in phylogenetic treespace: cortical landmark correspondence and descendant correspondence. The differences in end-results based on these mappings are highlighted to emphasize the importance of correspondence in tree-oriented data analysis. Representation of brain artery systems as points in phylogenetic treespace, a mathematical space developed in (Billera et al. Adv. Appl. Math 27:733–767, 2001), facilitates this analysis. The phylogenetic treespace is a rich setting for tree-oriented data analysis. The Fréchet sample mean or an approximation is reported. Multidimensional scaling is used to explore structure in the data set based on pairwise distances between data points. This analysis of MRA data shows a statistically significant effect of age and sex on brain artery structure. Variation in the proximity of brain arteries to the cortical surface results in strong statistical difference between sexes and statistically significant age effect. That particular observation is possible with cortical correspondence but did not show up in the descendant correspondence.

















Similar content being viewed by others
References
Asfari, B., Tron, R., Vidal, R.: On the convergence of gradient descent for finding the Riemannian center of mass. SIAM J. Control Optim. Accepted (2013). http://www.cis.jhu.edu/~bijan/
Aydin, B., Pataki, G., Wang, H., Bullitt, E., Marron, J.: A principal component analysis for trees. Ann. Appl. Stat. 3, 1597–1615 (2009)
Aydin, B., Pataki, G., Wang, H., Ladha, A., Bullitt, E., Marron, J.: Visualizing the structure of large trees. Electron. J. Stat. 5, 405–420 (2009)
Aylward, S., Bullit, E.: Initialization, noise, singularities, and scale in height ridge traversal for tubular object centerline extraction. IEEE Trans. Med. Imaging 21, 61–75 (2002)
Barden, D., Le, H., Owen, M.: Central limit theorems for fréchet means in the space of phylogenetic trees. Electron. J. Probab. (2013)
Bac̆àk, M.: A novel algorithm for computing the Fréchet mean in Hadamard spaces (2012). arXiv:1210.2145v1
Bhattacharya, R., Patrangenaru, V.: Large sample theory of intrinsic and extrinsic sample means on manifolds. Ann. Stat. (2003), 29 pp.
Billera, L., Holmes, S., Vogtmann, K.: Geometry of the space of phylogenetic trees. Adv. Appl. Math. 27, 733–767 (2001)
Bridson, M., Haefliger, A.: Metric Spaces of Non-positive Curvature. Springer, Berlin (1999)
Bullitt, E., Smith, J., Lin, W.: Designed database of MR brain images of healthy volunteers (2008)
Dryden, I., Mardia, K.: Statistical Shape Analysis. Wiley, New York (1998)
Felsenstein, J.: Inferring Phylogenies. Sinauer Associates (2004)
Feragen, A., Lo, P., de Bruijne, M., Nielson, M., Lauze, F.: Towards a theory of statistical tree-shape analysis. IEEE Trans. Pattern Anal. Mach. Intell. 20 (2012)
Fréchet, M.: Les élément aléatoires de nature quelconque dans un espace distancié. Ann. Inst. Henri Poincaré 10, 215–310 (1948)
Hillis, D., Heath, T., John, K.S.: Analysis and visualization of tree space. Syst. Biol. 54(3), 471–482 (2005)
Hotz, T., Huckemann, S., Le, H., Marron, J.S., Mattingly, J.C., Miller, E., Nolen, J., Owen, M., Patrangenaru, V., Skwerer, S.: Sticky central limit theorems on open books. arXiv:1202.4267 (2012)
Lee, P., Chang, H., Lin, C., Chiang, A., Ching, Y.: Constructing neuronal structure from 3d confocal microscopic images. J. Med. Biol. Eng. 29, 1–6 (2009)
Miller, E., Owen, M., Provan, J.: Averaging metric phylogenetic trees (2012). arXiv:1211.7046
Nye, T.: Principal components analysis in the space of phylogenetic trees. Ann. Stat. 39, 2243–2794 (2011)
Oguz, I., Cates, J., Fletcher, T., Whitaker, R., Cool, D., Aylward, S., Styner, M.: Cortical correspondence using entropy-based particle systems and local features. In: Biomedical Imaging: From Nano to Macro, ISBI 2008, 5th IEEE International Symposium on, pp. 1637–1640. IEEE, New York (2008)
Owen, M., Provan, S.: A fast algorithm for computing geodesic distances in tree space. Comput. Biol. Bioinform. 8, 2–13 (2011)
Shen, D., Shen, H., Bhamidi, S., Muñoz, Y.M., Kim, Y., Marron, J.: Functional data analysis of tree data objects. J. Comput. Graph. Stat., accepted (2014)
Sturm, K.: Probability measures on metric spaces of nonpositive curvature. In: Heat Kernels and Analysis on Manifolds, Graphs, and Metric Spaces. Contemp. Math., vol. 338, pp. 357–390 (2003). Lecture Notes from a quarter program on heat kernels, random walks, and analysis on manifolds and graphs
Wang, J.: Geometric Structure of High-Dimensional Data and Dimensionality Reduction. Springer, Berlin (2011)
Acknowledgements
The MR brain images form healthy volunteers used in this paper were collected and made available by the CASILab at The University of North Carolina at Chapel Hill and were distributed by the MIDAS Data Server at Kitware, Inc.
Author information
Authors and Affiliations
Corresponding author
Additional information
We are grateful to SAMSI, for sponsoring the authors’ visits to the Research Triangle and for facilitating net meetings, as part of the Analysis of Object Data program in 2010–2011, that helped advance this research. SH acknowledges support from DFG HU 1575/2-1 as well as the Niedersachsen Vorab of the Volkswagen Foundation. IO acknowledges support from UNC Neurodevelopment Disorders Research Center HD 03110. MO acknowledges the support of the Fields Institute VP acknowledges support from NSF grants DMS-0805977 and DMS-1106935.
Rights and permissions
About this article
Cite this article
Skwerer, S., Bullitt, E., Huckemann, S. et al. Tree-Oriented Analysis of Brain Artery Structure. J Math Imaging Vis 50, 126–143 (2014). https://doi.org/10.1007/s10851-013-0473-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10851-013-0473-0