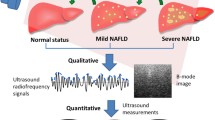
Abstract
Fatty Liver Disease (FLD) is caused by the deposition of fat in liver cells and leads to deadly diseases such as liver cancer. Several FLD detection and characterization systems using machine learning (ML) based on Support Vector Machines (SVM) have been applied. These ML systems utilize large number of ultrasonic grayscale features, pooling strategy for selecting the best features and several combinations of training/testing. As result, they are computationally intensive, slow and do not guarantee high performance due to mismatch between grayscale features and classifier type. This study proposes a reliable and fast Extreme Learning Machine (ELM)-based tissue characterization system (a class of Symtosis) for risk stratification of ultrasound liver images. ELM is used to train single layer feed forward neural network (SLFFNN). The input-to-hidden layer weights are randomly generated reducing computational cost. The only weights to be trained are hidden-to-output layer which is done in a single pass (without any iteration) making ELM faster than conventional ML methods. Adapting four types of K-fold cross-validation (K = 2, 3, 5 and 10) protocols on three kinds of data sizes: S0-original, S4-four splits, S8-sixty four splits (a total of 12 cases) and 46 types of grayscale features, we stratify the FLD US images using ELM and benchmark against SVM. Using the US liver database of 63 patients (27 normal/36 abnormal), our results demonstrate superior performance of ELM compared to SVM, for all cross-validation protocols (K2, K3, K5 and K10) and all types of US data sets (S0, S4, and S8) in terms of sensitivity, specificity, accuracy and area under the curve (AUC). Using the K10 cross-validation protocol on S8 data set, ELM showed an accuracy of 96.75% compared to 89.01% for SVM, and correspondingly, the AUC: 0.97 and 0.91, respectively. Further experiments also showed the mean reliability of 99% for ELM classifier, along with the mean speed improvement of 40% using ELM against SVM. We validated the symtosis system using two class biometric facial public data demonstrating an accuracy of 100%.










Similar content being viewed by others
Change history
07 December 2017
The original version of this article unfortunately contained a mistake. The family name of Rui Tato Marinho was incorrectly spelled as Marinhoe.
References
Saverymuttu, S.H., Joseph, A.E., and Maxwell, J.D., Ultrasound scanning in the detection of hepatic fibrosis and steatosis. Br Med J (Clin Res Ed). 292(6512):13–15, 1986.
Mohamed, W.S., Mostafa, A.M., Mohamed, K.M., and Serwah, A.H., The epidemiology of nonalcoholic fatty liver disease in adults by Clark, Jeanne M MD, MPH. J. Clin. Gastroenterol. 40:S5–S10, 2006.
Wieckowska, A., and Feldstein, A.E., Nonalcoholic fatty liver disease in the pediatric population: A review. Current opinion in pediatrics. 17(5):636–641, 2005.
Ratziu, V., Charlotte, F., Heurtier, A., Gombert, S., Giral, P., Bruckert, E., Grimaldi, A., and Capron, F., Thierry Poynard, and LIDO study group, sampling variability of liver biopsy in nonalcoholic fatty liver disease. Gastroenterology. 128(7):1898–1906, 2005.
Cheung, R.C., Complications of liver biopsy, gastrointestinal emergencies, gastrointestinal emergencies. In: Tham, T.C.K., Collins, J.S.A., and Soetikno, R. (Eds.). Blackwell, West Sussex, UK, pp. 72–79, 2009.
Saadeh, S., Younossi, Z.M., Remer, E.M., Gramlich, T., Ong, J.P., Hurley, M., Mullen, K.D., Cooper, J.N., and Sheridan, M.J., The utility of radiological imaging in nonalcoholic fatty liver disease. Gastroenterology. 123(3):745–750, 2009.
Wang, D., Fang, Y., Hu, B., Cao, H., B-scan image feature extraction of fatty liver. In Internet Computing for Science and Engineering (ICICSE), 2012 Sixth International Conference. (2012) 188–192.
Yajima, Y., Ohta, K., Narui, T., Abe, R., Suzuki, H., and Ohtsuki, M., Ultrasonographical diagnosis of fatty liver: Significance of the liver-kidney contrast. Tohoku. J. Exp. Med. 139(1):43–50, 1983.
Mathiesen, U.L., Franzen, L.E., Aselius, H., Resjö, M., Jacobsson, L., Foberg, U., Frydén, A., and Bodemar, G., Increased liver echogenicity at ultrasound examination reflects degree of steatosis but not of fibrosis in asymptomatic patients with mild/moderate abnormalities of liver transaminases. Dig. Liver Dis. 34(7):516–522, 2002.
Mendler, M.H., Bouillet, P., Le Sidaner, A., Lavoine, E., Labrousse, F., Sautereau, D., and Pillegand, B., Dual-energy CT in the diagnosis and quantification of fatty liver: Limited clinical value in comparison to ultrasound scan and single-energy CT, with special reference to iron overload. J. Hepatol. 28(5):785–794, 1998.
Acharya, U., and Rajendra, J., Suri, data mining framework for fatty liver disease classification in ultrasound: A hybrid feature extraction paradigm. Med. Phys. 39(7):4255–4264, 2012.
Shensa, M.J., The discrete wavelet transform: The discrete wavelet transform: Wedding the atrous and Mallat algorithms. IEEE. Trans. Signal Process. 40(10, 1992):2464–2482.
Bruce, L.M., Koger, C.H., and Li, J., Dimensionality reduction of hyperspectral data using discrete wavelet transform feature extraction. IEEE. Trans. Geosci. Remote. Sens. 40(10):2331–2338, 2002.
Manjunath, B.S., and Ma, W.Y., Texture features for browsing and retrieval of image data. IEEE. Trans. Pattern. Anal. Mach. Intellig. 18(8):837–842, 1996.
Ishibuchi, H., Nakashima, T., and Murata, T., Performance evaluation of fuzzy classifier systems for multidimensional pattern classification problems. IEEE. Trans. Syst. Man. Cybern. Part B (Cybern.). 29(5):601–618, 1999.
Kuncheva L., Fuzzy classifier design, Springer Science & Business Media; 2000 Apr 26.
Uebele, V., Abe, S., and Lan, M.-S., A neural-network-based fuzzy classifier. IEEE. Trans. Syst. Man. Cybern. 25(2):353–361, 1995.
Antonini, M., Barlaud, M., Mathieu, P., and Daubechies, I., Image coding using wavelet transform. IEEE. Trans. Image. Process. 1(2):205–220, 1992.
Subramanya, M.B., Kumar, V., Mukherjee, S., and Saini, M., A CAD system for B-mode fatty liver ultrasound images using texture features. J. Med. Eng. Technol. 39(2):123–130, 2015.
Ma, H.Y., Zhou, Z., Wu, S., Wan, Y.L., and Tsui, P.H., A computer-aided diagnosis scheme for detection of fatty liver in vivo based on ultrasound kurtosis imaging. J. Med. Syst. 40(1):33, 2016.
Saba, L., Dey, N., Ashour, A.S., Samanta, S., Nath, S.S., Chakraborty, S., Sanches, J., Kumar, D., Marinho, R., and Suri, J.S., Automated stratification of liver disease in ultrasound: An online accurate feature classification paradigm. Comput. Methods. Programs. Biomed. 130:118–134, 2016.
Vapnik VN, An overview of statistical learning theory, IEEE transactions on neural networks (10) (1999) 988–999.
Huang, G.-B., and Zhu, Q.-Y., Chee-Kheong Sie extreme learning machine: Theory and applications. Neurocomputing. 70(1):489–501, 2006.
Huang, G.-B., Zhu, Q.-Y., and Siew, C.-K., Extreme learning machine: A new learning scheme of feedforward neural networks, in neural networks, 2004. Proceedings. IEEE. Int. Joint. Conference. 2(2004):985–990, 2004.
Rao, C.R., and Mitra, S.K., Generalized inverse of matrices and its applications. Wiley, New York, 1971.
Qayyum, A., MR spectroscopy of the liver: Principles and clinical applications. Radiographics. 29(6):1653–1664, 2009.
Kadah, Y.M., Farag, A.A., Zurada, J.M., Badawi, A.M., and Youssef, A.M., Classification algorithms for quantitative tissue characterization of diffuse liver disease from ultrasound images. IEEE Trans. Med. Imaging. 15(4):466–478, 1996.
Chorowski, J., Wang, J., and Zurada, J.M., Review and performance comparison of SVM-and ELM-based classifiers. Neurocomputing. 128:507–516, 2014.
Acharya, U.R., Sree, S.V., Krishnan, M.M., Molinari, F., ZieleŸnik, W., Bardales, R.H., Witkowska, A., and Suri, J.S., Computer-aided diagnostic system for detection of Hashimoto thyroiditis on ultrasound images from a polish population. J. Ultrasound. Med. 33:245–253, 2014.
Mohanty, A.K., Beberta, S., and Lenka, S.K., Classifying benign and malignant mass using GLCM and GLRLM based texture features from mammogram. Int. J. Eng. Res. Appl. 1(3):687–693, 2011.
Mohanaiah, P., and Sathyanarayana, L., GuruKumar, image texture feature extraction using GLCM approach. Int J. Sci. Res. Publ. 3(5):1, 2013.
Herman, P., Comparative analysis of spectral approaches to feature extraction for EEG-based motor imagery classification. IEEE Trans. Neural Syst. Rehabil. Eng. 16(4):317–326, 2008.
Anuradha, K., Statistical feature extraction to classify oral cancers. J.Glob. Res. Comput. Sci. 4(2):8–12, 2013.
Barbu, T., Gabor filter-based face recognition technique. Proc. Rom. Acad. 11(3):277–283, 2010.
MacAusland, Ross, The Moore-Penrose Inverse and Least Squares, Math 420. Advanced Topics in Linear Algebra. (2014).
Mirmehdi M, Handbook of texture analysis, Imperial College Press. 2008.
Acharya, U.R., Mookiah, M.R., Sree, S.V., Afonso, D., Sanches, J., Shafique, S., Nicolaides, A., Pedro, L.M., e Fernandes, J.F., and Suri, J.S., Atherosclerotic plaque tissue characterization in 2D ultrasound longitudinal carotid scans for automated classification: A paradigm for stroke risk assessment. Med. Biol. Eng. Comput. 51(5):513–523, 2013.
Acharya, R.U., Faust, O., Alvin, A.P., Sree, S.V., Molinari, F., Saba, L., Nicolaides, A., and Suri, J.S., Symptomatic vs. asymptomatic plaque classification in carotid ultrasound. J. Med. Syst. 36(3):1861–1871, 2012.
Acharya, U.R., Faust, O., Sree, S.V., Molinari, F., Saba, L., Nicolaides, A., and Suri, J.S., An accurate and generalized approach to plaque characterization in 346 carotid ultrasound scans. IEEE. Trans. Instrum. Meas. 61(4):1045–1053, 2012.
Shrivastava, V.K., Computer-aided diagnosis of psoriasis skin images with HOS, texture and color features: A first comparative study of its kind. Comput Meth. Programs Biomed. 126:98–109, 2016.
Shrivastava, V.K., Londhe, N.D., Sonawane, R.S., and Suri, J.S., Reliable and accurate psoriasis disease classification in dermatology images using comprehensive feature space in machine learning paradigm. Expert Syst. Appl. 42:6184–6195, 2015.
Douali, N., Abdennour, M., Sasso, M., Miette, V., Tordjman, J., Bedossa, P., Veyrie, N., Poitou, C., Aron-Wisnewsky, J., Clément, K. and Jaulent, M.C. Noninvasive diagnosis of nonalcoholic steatohepatitis disease based on clinical decision support system. MedInfo. 192:1178, 2013.
Vanderbeck, S., Bockhorst, J., Komorowski, R., Kleiner, D.E., and Gawrieh, S., Automatic classification of white regions in liver biopsies by supervised machine learning. Hum. Pathol. 45(4):785–792, 2014.
Liu, X., Song, J.L., Wang, S.H., Zhao, J.W., and Chen, Y.Q., Learning to diagnose cirrhosis with liver capsule guided ultrasound image classification. Sensors. 17(1):149, 2017.
LeCun, Y., Bengio, Y., and Hinton, G. Deep learning. Nature. 521(7553):436–444, 2015.
Huang, G.-B., Extreme learning machine for regression and multiclass classification. IEEE. Trans. Syst. Man. Cybern. Part B (Cybern). 42(2):513–529, 2012.
Zhu, Q.-Y., Evolutionary extreme learning machine. Pattern. Recogn. 38(10):1759–1763, 2005.
Li Mao, Lidong Zhang, Xingyang Liu, Chaofeng Li, Hong Yang, Improved Extreme Learning Machine and Its Application in Image Quality Assessment. Mathematical Problems in Engineering. (2014).
Tang, J., Deng, C., and Huang, G.-B., Extreme learning machine for multilayer perceptron. IEEE. Trans. Neural Netw. Learn. Syst. 27(4):809–821, 2016.
Demuth, H.B., Beale, M.H., De Jess, O. and Hagan, M.T., Neural network design, Martin Hagan, 2014.
El-Baz A, Suri JS, Big Data in Medical Imaging, CRC Press, 2018 (to appear).
El-Baz AS, Saba L, Suri JS. Abdomen and thoracic imaging. Springer, 2014.
El-Baz A, Gimel’farb G, Suri JS, Stochastic modeling for medical image analysis, CRC Press, 2015.
Esses SJ, Lu X, Zhao T, Shanbhogue K, Dane B, Bruno M, Chandarana H. Automated image quality evaluation of T2-weighted liver MRI utilizing deep learning architecture, Journal of Magnetic Resonance Imaging, (2017).
Acknowledgements
The authors of National Institute of Technology Goa, India would like to acknowledge Ministry of Human Resource department, Government of India and MediaLab Asia, Ministry of Electronics and Information Technology, Government of India for their kind support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
None of the authors have any conflict of interest.
Ethical Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Data was collected after IRB approval by Instituto Superior Tecnico (IST), University of Lisbon, Portugal and written informed consent provided by all the patients.
Additional information
This article is part of the Topical Collection on Image & Signal Processing
A correction to this article is available online at https://doi.org/10.1007/s10916-017-0862-9.
Appendices
Appendix A: ELM mathematical framework and solution
Let there be H hidden layer neurons and N output neurons. Let each training sample of P images be denoted as (a j , l j ), where each input image denoted as a j = [a j1, a j2, …, a jm ]T ∈ R m and each output label is denoted as l j = [l j1, l j2, …, l jN ]T ∈ R N. The output vector is denoted as L = [l 1, l 2, …, l j , …l P ]T. Further, the P images and their ground truth labels are divided into two parts for training and testing which can be represented by \( {\boldsymbol{P}}_{trg}=\left({\boldsymbol{a}}_j^{trg},{\boldsymbol{l}}_j^{trg}\right) \) for training and \( {\boldsymbol{P}}_{tst}=\left({\boldsymbol{a}}_k^{tst},{\boldsymbol{l}}_k^{tst}\right) \). Similarly, output vector L is also divided into two sets L trg and L tst . Each input layer neuron is connected to all hidden layer neurons. Let each hidden layer weight be denoted as a vector w i = [w 1i , w 2i , …, w mi ]T. Each of the connections or weights from hidden-to-output layer are denoted as δ i = [δ i1, δ i2, …, δ iN ]T connecting i th hidden node to the output nodes. A standard SLFFNN can be modeled as given by:
where, g is the activation function and b is the bias. This equation is written in more compact form which is given by:
where,
\( \boldsymbol{\delta} ={\left[\begin{array}{c}\hfill {\delta}_1^T\hfill \\ {}\hfill \begin{array}{c}\hfill \vdots \hfill \\ {}\hfill {\delta}_i^T\hfill \\ {}\hfill \vdots \hfill \end{array}\hfill \\ {}\hfill {\delta}_H^T\hfill \end{array}\right]}_{H\times N}\mathrm{and}\ {\boldsymbol{L}}_{trg}={\left[\begin{array}{c}\hfill {\boldsymbol{l}}_1^{trg}\hfill \\ {}\hfill \begin{array}{c}\hfill \vdots \hfill \\ {}\hfill {\boldsymbol{l}}_{\boldsymbol{j}}^{trg}\hfill \\ {}\hfill \vdots \hfill \end{array}\hfill \\ {}\hfill {\boldsymbol{l}}_{\#{\boldsymbol{P}}_{trg}}^{trg}\hfill \end{array}\right]}_{\#{\boldsymbol{P}}_{\boldsymbol{trg}}\times N}.\mathrm{The}\ \mathrm{cost}\ \mathrm{function}\ \mathrm{of}\ \mathrm{ELM}\ \mathrm{is}\ \mathrm{given}\ \mathrm{as}: \)
The objective is to find the minimum δ which minimizes the cost function E. By using Eq. 5, the Eq. 6 can also be written as:
where \( \hat{\boldsymbol{\delta}} \) is the least squares solution of the Qδ = L trg . If the number of hidden nodes is equal to the number of training samples (#P trg = H), the matrix Q is square and invertible. Therefore, with random weights w i and bias b i the training samples can be approximated with zero error. However, in maximum cases, number of training samples is larger than number of hidden nodes. So, the smallest norm least squares solution of the linear system is given by:
Where, Q † is the Moore–Penrose [35] generalized inverse of matrix Q. Thus the smallest training error can be reached by:
The trained hidden-to-output layer weights are then used in Testing-phase as shown in Fig. 5, to test the performance of the Symtosis model using test dataset P tst .
Appendix B: Support vector machine
Support Vector Machine (SVM) is a kernel based classification technique based on the maximum margin classifier. It transforms the original input data to high-dimensional feature space and tries to find the hyper-plane which maximizes the distance between data points of distinct classes. We consider a binary classification task with the training dataset designated as {(a i , L i ), i = 1, 2, …, l} where a i є R q is the input data for i th training sample and L i є [−1, +1] are the equivalent target values, l designates total number of samples and q is the input space dimension. The SVM model can be represented in feature space by following equation:

where, ℵ(x) represents kernel function, b represents bias and  is a weight vector which is normal to the hyper-plane. The decision rule is mathematically represented in the Eq. (11):
is a weight vector which is normal to the hyper-plane. The decision rule is mathematically represented in the Eq. (11):

The non-linear kernel function finds the maximum margin hyper-plane,  between the classes in a feature space. To find optimal hyper-plane, the Eq. (12) is minimized subject to Eqs. (10) and (11).
between the classes in a feature space. To find optimal hyper-plane, the Eq. (12) is minimized subject to Eqs. (10) and (11).

where, ϑ represents the trade-off between error and margin and ξ is a slack variable. By using Lagrangian multipliers (α) in dual form, the Eq. (12) can be transformed into following optimization problem:
The final decision function is given by following equation:
The parameters ωand γ define the separating hyperplane. The most general kernel functions are:
Where, deg. is the degree of kernel in Eq. 18 and (.) denotes the dot product.
Appendix C: Feature extraction
Haralick texture (GLCM)
GLCM calculates the following features shown Table 7 from the co-occurrence matrix calculated from the image.
Run length texture
GRLM feature extraction algorithm calculate features from the run length matrix as shown in Table 8.
Appendix D: Symbols table
Appendix E: Results of ELM/SVM classifier for S4 and S8 dataset
Appendix F: Scientific validation
Scientific validation is always an integrated component of the system design. For validation, one needs to run another set of liver data sets whose results are known a priori. Since such a clinical data is hard to obtain, we use facial biometric data set to test the classification accuracy. We do acknowledge Dr. Libor Spacek of Department of Computer Science, University of Essex for providing data on biometric facial dataset, namely Face94 [42]. This dataset consisting of male and female faces was experimented for validation using ELM/SVM.
Face94 data set
We have conducted experiments to validate our results using Face94 data set. The Face94 data set consists 153 individual images with various expressions and poses seated at a fixed distance from camera. There are 2 classes, male and female and total number of images are 2660, of which 2260 are male images and 400 are female images. A subset of images is given in Fig. 10. Cross-validation protocol (K2, K3, K5 and K10) is also performed to check generalization. The validation results are shown in Table 12. It is seen ELM gives 100% accuracy across all cross-validation protocols.
Rights and permissions
About this article
Cite this article
Kuppili, V., Biswas, M., Sreekumar, A. et al. Extreme Learning Machine Framework for Risk Stratification of Fatty Liver Disease Using Ultrasound Tissue Characterization. J Med Syst 41, 152 (2017). https://doi.org/10.1007/s10916-017-0797-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10916-017-0797-1